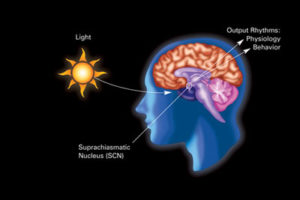
 For many years there was a consensus that most organisms have a circadian clock. In humans it was considered to be directed centrally by the master clock in the brain region suprachiasmatic nucleus (SCN). This clock appears to be involved in directing essential physiological processes throughout the body including secretion of hormones, metabolism, energy, and of course, sleep and wakefulness. It was not at all clear how this central clock works and how it communicates with so many different cellular physiological functions.
For many years there was a consensus that most organisms have a circadian clock. In humans it was considered to be directed centrally by the master clock in the brain region suprachiasmatic nucleus (SCN). This clock appears to be involved in directing essential physiological processes throughout the body including secretion of hormones, metabolism, energy, and of course, sleep and wakefulness. It was not at all clear how this central clock works and how it communicates with so many different cellular physiological functions.
Recently, it was a shock to the scientific community, when it was discovered that every cell has a fully functional clock, including microbes. The question now has become how all these cells interact together—how they coordinate their clocks, their genes, and their activity.
One possible way that cells’ individual clocks influence other cells and organs was discovered. A very recent study found that in the human gut, microbes living near lining cells move in a particular timed pattern throughout the day. By the clock, they move micrometers to the left, then the right, and then back again to the starting point. This simple movement appears to influence the circadian rhythm in the animal because the gut lining cells are having different interactions with various microbes in a specific rhythm throughout the day.
 This is one of many new findings related to the existence of clocks in each cell. Many previous posts have demonstrated that most of life’s activity occurs because of cellular conversations. This post will discuss how cells might communicate using their clocks, and in particular how individual cells and tissue cells can coordinate with central clocks in animals. The next post will discuss how clocks can interact with complex metabolic cycles in each organ.
This is one of many new findings related to the existence of clocks in each cell. Many previous posts have demonstrated that most of life’s activity occurs because of cellular conversations. This post will discuss how cells might communicate using their clocks, and in particular how individual cells and tissue cells can coordinate with central clocks in animals. The next post will discuss how clocks can interact with complex metabolic cycles in each organ.
First, a brief summary of a previous post on the influence of light. It noted that there is one central clock (SCN in the hypothalamus) and many peripheral clocks in bodily tissues that interact with the environment and the central clock. Peripheral clocks are in esophagus, lungs, liver, pancreas, spleen, thymus, skin, and prostate. Liver cells respond to food, not light. Many parts of the brain, including areas critical for mood and cognition have their own peripheral clocks that interact with the central system.
 External light can reset some aspects of the clock, including the cycle of core body temperature, brain waves, and hormone production. In the migration of butterflies, the clock determines both the sun compass for bright days and magnetic compass for cloudy days.
External light can reset some aspects of the clock, including the cycle of core body temperature, brain waves, and hormone production. In the migration of butterflies, the clock determines both the sun compass for bright days and magnetic compass for cloudy days.
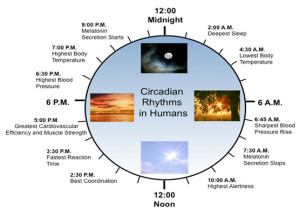
A summary of the effects of circadian clock on human metabolism:
- Early morning – stops melatonin, increases blood pressure, stimulates bowel movement, increases testosterone, most alert, best coordination
- Afternoon – fastest reaction time, best cardiovascular efficiency, most muscle strength,
- Evening – highest blood pressure and body temperature
- Night – suppress bowel movements, more melatonin, sleep
Independent Cellular Clocks
Inside of almost every cell there are clocks based on a series of genetic networks with loops, both feed forward and backward. These loops involve many different layers of genetic regulation including DNA, messenger RNA, and proteins. Also, epigenetic tags to DNA, RNA, and proteins are part of the loops.
The network of genes that produce timed loops for the internal clock of the cell has a strange and hard to remember name. It is called TTFL for transcriptional-translational feedback loop. Transcription, we remember is the process of making messenger RNA from pieces of the DNA code. In fact, this is a complex process with editing out of introns (pieces of DNA that aren’t part of the code), placing exons together (pieces left after taking out introns), and then shuffling these pieces in different combinations (called alternative splicing). The next process is called translation where a protein is made from a piece of messenger RNA in the ribosome. This protein can then act upon the DNA or RNA creating a loop.
What is quite remarkable is that these independent clocks in individual cells using genetic networks somehow communicate with tissues and the outside world to sync with other clocks based on other genetic networks.
 In all creatures in nature that react to light, scientists are finding 24 hour clocks. Almost 200 years ago this was first observed in the Mimosa tree where leaves open and close each day at particular times. The first known organism with a clock is thought to be cyanobacteria 2 billion years ago. In these bacteria the clock appeared to be related to the time that photosynthesis occurred. One factor that required a clock is timing the very difficult process of repairing damaged DNA during sunlight. Another was how not to waste time with unsuccessful search for energy and food.
In all creatures in nature that react to light, scientists are finding 24 hour clocks. Almost 200 years ago this was first observed in the Mimosa tree where leaves open and close each day at particular times. The first known organism with a clock is thought to be cyanobacteria 2 billion years ago. In these bacteria the clock appeared to be related to the time that photosynthesis occurred. One factor that required a clock is timing the very difficult process of repairing damaged DNA during sunlight. Another was how not to waste time with unsuccessful search for energy and food.
The clock allows cells and organisms to plan ahead to maximize their efforts. This is true for both individual bacteria cells and humans. It is remarkable that a genetic network can help plan ahead for bacteria.
Clock Made of Genes and Proteins that Influence Genes
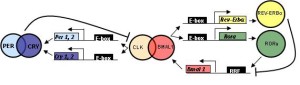
While clocks were first noticed two centuries ago, only 12 years ago, specific genes were found to create networks and loops to produce the clocks. These made protein factors that landed on sections of DNA to stimulate or repress particular sections (transcription factors). The first clock transcription factor discovered was PER (for “period”) and this was noted to have a 24-hour cycle of activity. PER is a factor that represses critical factors that activate a feedback loop. PER represses the production of transcription factors CLOCK and BMAL1. BMAL1 is also known as NPAS2. Also, PER works with another repressor called CRY (for “crytochrome”). Another repressor that works after PER is called Rev-erb.
First Per gene produces PER protein (by transcription of DNA to messenger RNA to a protein). It is then moved to a place where it can repress a part of the cycle.
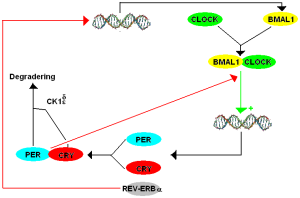
What is unusual is that these components of a 24 hour cycles respond as other molecules because of changes in chemicals near them. All of these transcription factors mentioned (PER, CLOCK, BMAL1) have a section of the molecule that is called PAS. PAS is known to be part of many different transcription factors related to response to oxygen and metabolism. The other factor Rev-erb is one of many receptors in the nucleus (nuclear receptors NR) that respond to many metabolic molecules, hormones, and other chemical.
As well as the transcription factors that start and stop DNA functions, another level of regulation of the cycle is from altering signals, or tags, on the proteins themselves. An example occurs with PER, where a phosphate tag molecule is added, which alters their chemical properties at different temperatures. Another is adding a ubiquitin tag to CRY, which alters its function. The adding of particular tags such as phosphate and ubiquitin are vital ways that the cycles operate.
While some of these examples are known, the basic mechanisms are still incredibly complex and not well understood. An example of what we don’t know is how everything changes if the environment suddenly changes. One piece of information that has been discovered is that several sleep disorders appear to be based on a mutation in some of these key molecules.
Many Kinds of Cells and Many Kinds of Clocks
 The basic clock mechanism appears to be very similar in all cells, such as the transcription factors noted and their feedback loops. Of course, these transcription factors work on DNA and therefore the DNA has to be accessible. DNA is normally kept wound up on a protein spool and not able to be used until it is unwound or opened. For DNA to be used, the histone spools that protect strands of DNA must be opened.
The basic clock mechanism appears to be very similar in all cells, such as the transcription factors noted and their feedback loops. Of course, these transcription factors work on DNA and therefore the DNA has to be accessible. DNA is normally kept wound up on a protein spool and not able to be used until it is unwound or opened. For DNA to be used, the histone spools that protect strands of DNA must be opened.
Special tags on the histone spools keep a region of DNA closed or open. Both transcription factors and special tags on the histones allow the DNA region to open or not. Study of RNA has found that as many as 10% of all known RNAs are part of this cycling. What is surprising is that the types of RNA that are cycling are different in each bodily tissue. Many of the genes are also specific to the type of tissue and therefore are different from the central clock mechanism. Recently, it has been found that transcription factors such as enhancers are also different in each type of cell and this occurs in the fetus.
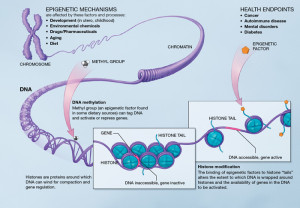 These different mechanisms in each tissue are now known to include all of genetic regulatory levels—protein transcription factors, RNAs, and specific enzymes that place and remove tags on histones and alter the structures of the spools themselves. One histone tag and related enzyme is particularly tied to the clock rhythms in the liver—acetyl molecules placed on by enzymes called acetyl histone transferases (HATs) and taken off by deacetylase (HDACs). Another tag using methyl groups is part of this same process.
These different mechanisms in each tissue are now known to include all of genetic regulatory levels—protein transcription factors, RNAs, and specific enzymes that place and remove tags on histones and alter the structures of the spools themselves. One histone tag and related enzyme is particularly tied to the clock rhythms in the liver—acetyl molecules placed on by enzymes called acetyl histone transferases (HATs) and taken off by deacetylase (HDACs). Another tag using methyl groups is part of this same process.
While the processes are vastly complex, it appears that the original fundamental consistent core clock processes open up the relevant DNA and then this is used in variable ways in different tissues.
There are new levels of complexity being discovered. An example is interactions with the fundamental CRY and a receptor for steroids (glucocorticoid receptor or GR). CRY can affect many different regions of the DNA and helps regulate how GR is activated and repressed. In different tissues, these regions operating can be in very different and distant places
Other very specific ways that the core clock proteins are altered are from particular vital metabolic molecules—acetyl CoA, S-adenosyl methionine (SAM) and the vital energy particle nicotinamide adenine dinucleotide (NAD+). (The next post covers this relationship for these as well as cholesterol and heme). These are found to involve the enzymes mentioned that alter the tags on histones related to these metabolites. What makes this very complex is all the central clock proteins can be altered by tags on themselves or the histones.
Great Complexity Discovered
 Regulation of the many interacting clocks for particular molecules is vastly complex with regulation at all levels—specific DNA repression and activation, alteration of transcription factors, tags on histone to open pieces of DNA, alterations of RNAs, and tags on proteins.
Regulation of the many interacting clocks for particular molecules is vastly complex with regulation at all levels—specific DNA repression and activation, alteration of transcription factors, tags on histone to open pieces of DNA, alterations of RNAs, and tags on proteins.
New discoveries that affect rhythms are alterations in proteins that form oscillations, and variations in a particular region of messenger RNA called polyadenylation. Alterations molecular rhythms occurs along with loops already described with changing transcription factors, tags, and RNAs. Polyadenylation are sections of messenger RNA that can be altered, that change the way messenger RNA is transported out of the nucleus.
There are many other cellular rhythms. When molecules are oxidized (reacting with oxygen) the specialized proteins (called redox) all have daily rhythms. It is possible that the cycle of cellular reproduction is tied to cellular clocks, synchronized through cycles of ability to find food and scarcity.
Organization of Individual Clocks in Tissues
 Coordination with the central mechanism is different in each type of cell. Individual cell clocks are coordinated with the central clock in a part of the brain called the suprachiasmatic nucleus (SCN). The SCN is in the hypothalamus, which is the main regulator of all bodily functions. It is highly connected with hormones, sleeping and waking, the sympathetic and parasympathetic control of internal organs, and eating and hunger.
Coordination with the central mechanism is different in each type of cell. Individual cell clocks are coordinated with the central clock in a part of the brain called the suprachiasmatic nucleus (SCN). The SCN is in the hypothalamus, which is the main regulator of all bodily functions. It is highly connected with hormones, sleeping and waking, the sympathetic and parasympathetic control of internal organs, and eating and hunger.
Light appears to be the main stimulus from the environment for the clock and rhythms controlling behavior. Light stimulates receptors with the molecule melanopsin and is highly connected to the SCN. Neurons in the SCN are altered through transcription factor called CREB. Alterations in the levels of some other enzymes related to redox alter these neurons as well. SCN signals centers for sleep and wakening.
The extremely complex mechanisms of sleep have not been worked out, but recent work shows significant role of a particular molecular system where small peptides interact and coordinate both the clock and the energy metabolic system. These neuropeptides are hypocretin and orexin. Changes in these cause the sleep disorder narcolepsy and to obesity, and other metabolic diseases.
 There are many ways that rhythms are produced that are not understood or just barely understood. Previous posts have described some poorly understood electric mechanisms in neurons called coupling. (see post on Electricity and the Brain). This coupling occurs in the neurons of the SCN that are related to the influences of clocks in the SCN and the tissue cells. Most discoveries related to this is in the liver producing schedules for eating and sleeping.
There are many ways that rhythms are produced that are not understood or just barely understood. Previous posts have described some poorly understood electric mechanisms in neurons called coupling. (see post on Electricity and the Brain). This coupling occurs in the neurons of the SCN that are related to the influences of clocks in the SCN and the tissue cells. Most discoveries related to this is in the liver producing schedules for eating and sleeping.
- There is evidence of unknown mechanisms that coordinate temperature through cycles and rhythms of RNA splicing when making messenger RNA.
- There is evidence of an unknown blood factor that triggers the tissue cell clocks.
- Another mechanism of communication occurs when one organ is ill and signals to other organs to alter their function. This has been described in lung disease that alters metabolic energy particles (NAD+) in other tissues.
- Cancer cells are able to alter clocks in peripheral cells. In fact, altering this circadian coordination can increase tumors.
Health and Disease
 Coordinating clocks is one way that the brain is able to plan for events in the body related to the environment and alter behavior. This relationship occurs through triggering gene networks. When the tissue cannot keep up with the coordination then it can become ill.
Coordinating clocks is one way that the brain is able to plan for events in the body related to the environment and alter behavior. This relationship occurs through triggering gene networks. When the tissue cannot keep up with the coordination then it can become ill.
In human history before electricity and lights and before access to food at all times, clocks were set by the environment. Now, food is always available and there are constant lights—blue computer lights from the computer and cell phone screens. Also, there is rapid travel from time zones. Another current factor is shift work at night, and work with altered schedules including night work.
In animal research, disruption of these factors produces changes in metabolism and many different medical issues including cancer. In insects and mice there is early aging, which includes more DNA breaks without repair causing more chance of cancer. Mice with mutant PER have more cancers. The opposite can occur, also, alteration of CRY can decrease risk. There are so many complex cycles that results can go in many directions.
One very complex and vital function in all cells is repairing DNA damage. This is so important that decreases in this ability have been considered to be a cause of aging. This involves many different protein enzymes. Because most DNA damage in the distant past was through ultraviolet light, circadian rhythms now have the maximum amount of these proteins when there is the most sunlight. Another source of damage is oxidation and related enzymes are also at the peak time of the photosynthesis process—the ancient source of oxidation problems. The relation to modern lifestyle factors is still too complex to make simple judgements.
Glucose metabolism is highly complex and highly regulated. Altered glucose metabolism has been associated with aging. This has been impacted by night shift work and jet lag. As mentioned melatonin is related to the amount of insulin. Studies have shown that the clocks in the liver and the pancreas are not the same and are in fact opposite. This is coordinated through complex processes. (More about clocks and metabolism in the next post).
Each Cell Has A Clock
 It has not been known how the central circadian clock can be translated into so many varied physiological functions. The recent discoveries that each cell has its own clock in addition to central larger clocks in many organs has opened the new question as to how all these clocks relate to each other. The genetic, RNA and protein loops somehow exist to anticipate the cycle of light and eating.
It has not been known how the central circadian clock can be translated into so many varied physiological functions. The recent discoveries that each cell has its own clock in addition to central larger clocks in many organs has opened the new question as to how all these clocks relate to each other. The genetic, RNA and protein loops somehow exist to anticipate the cycle of light and eating.
Research shows that cells have constant back and forth dialogue about all physiological functions. Therefore, it is not really surprising to find elaborate communication among all cells about timing related to each cell’s clock. This adds to the flood of new cellular signals being discovered every day. The next post discusses the new frontier of individual cell clocks in each tissue and how they relate to metabolism.