 Two hundred and fifty thousand cells a second migrating into place in the fetal brain. Finally, a trillion neurons are in place, each attempting to respond. Neurons capturing the incoming flood of sensory data, and those responding survive the pruning. The rest of the 900 billion cells are gone, systematically broken up, materials recycled, with no inflammation.
Two hundred and fifty thousand cells a second migrating into place in the fetal brain. Finally, a trillion neurons are in place, each attempting to respond. Neurons capturing the incoming flood of sensory data, and those responding survive the pruning. The rest of the 900 billion cells are gone, systematically broken up, materials recycled, with no inflammation.
A thousand connections on each of the 100 billion remaining neurons equals 100 trillion synapses, an almost incomprehensive amount. At the current rate of progress it will take thousands of years to diagram this many neuronal connections. In addition, each of these trillions of synapses has thousands of large interacting proteins inside and between connecting neurons.
A mental event instantly triggers these neurons with a vast cascade of molecular machinery, signaling to the nucleus, regulating the genetic production of necessary molecules. How complex is the regulation of these molecular cascades?
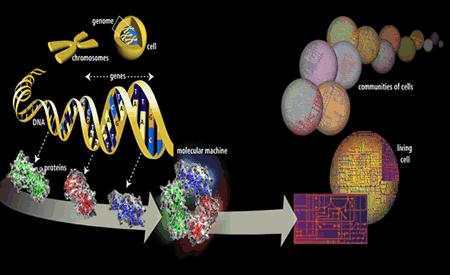 The previous post, part I of this series on the mind affecting molecular genetics, attempted to paint a picture of the complex regulation of the genetic elements in the cell including many protein transcription factors and a host of newly discovered small regulatory RNA molecules. It described some of the understanding of the genetic processes just prior to the recent explosion of data from ENCODE. A previous post also attempted to describe the complexity of genetic processes with the illustration of DNA self-editing. The cellular genetic editing involves complex analysis, cutting, splicing, and joining of each molecule that is produced before it is released into the action of the cell. This post begins the discussion of the new data from ENCODE.
The previous post, part I of this series on the mind affecting molecular genetics, attempted to paint a picture of the complex regulation of the genetic elements in the cell including many protein transcription factors and a host of newly discovered small regulatory RNA molecules. It described some of the understanding of the genetic processes just prior to the recent explosion of data from ENCODE. A previous post also attempted to describe the complexity of genetic processes with the illustration of DNA self-editing. The cellular genetic editing involves complex analysis, cutting, splicing, and joining of each molecule that is produced before it is released into the action of the cell. This post begins the discussion of the new data from ENCODE.
The Encyclopedia of DNA Elements – ENCODE
Up until recently it was thought that only about 2% of the DNA coded for proteins, (DNA regions that code for proteins have been called the “gene”) with some additional DNA to serve as sites where the enhancers and repressors are triggered. These DNA regulating regions, which sit near the gene, either stimulate or suppress the creation of proteins. The enhancers and repressors, as well as activators and promoters, trigger an effect when a protein (called a transcription factor) attaches to a particular location on the DNA.
This simple conception of the gene had to change when it was discovered that the DNA of one gene was not continuous but rather in pieces with irrelevant sections of DNA in between the relevant coding sections. The relevant pieces of the gene, which are used to code for specific proteins, are called exons, and the pieces that lie between exons are called introns and were considered junk.  The RNA pieces made from all of the exons of a particular gene are then spliced together to make the messenger RNA that finally manufactures a protein. It was known before ENCODE that the pieces could be spliced together in different ways making many more proteins than the 20,000 genes identified in the Genome Project ten years ago.
The RNA pieces made from all of the exons of a particular gene are then spliced together to make the messenger RNA that finally manufactures a protein. It was known before ENCODE that the pieces could be spliced together in different ways making many more proteins than the 20,000 genes identified in the Genome Project ten years ago.
For the past decade since the Genome Project, ENCODE has been analyzing the 98% of alleged “junk” DNA, that is, the DNA that is not part of the coding sections, which were not considered relevant. This “junk” was considered to be refuse from the very messy process of evolution. The Encyclopedia of DNA elements, or ENCODE, two weeks ago simultaneously published 30 seminal articles with the conclusions of hundreds of research projects around the world. ENCODE has been the largest international cooperative science venture in history.
Re-Definition of the Gene
 With the new data, it appears that the great complexity of human genetics is not in the number of genes (the rather small number of 20,000), but rather the amount of regulation of the genes. In fact, it was found that the neat description of a gene might have to be altered.
With the new data, it appears that the great complexity of human genetics is not in the number of genes (the rather small number of 20,000), but rather the amount of regulation of the genes. In fact, it was found that the neat description of a gene might have to be altered.
One of the striking findings of ENCODE is that many of the exons, (the little pieces of discrete DNA that together make up a “gene”) operate in multiple different “genes”. RNA will need greatly increased direction to splice together exons, the coding pieces of DNA, from multiple different locations and genes. Previously, the conception was that RNA merely had to cut out the introns of a section of DNA with many exons already in order.
With this change the definition of the gene can not continue to be specific exons involved in making one protein. Some ENCODE analysts are now calling for an entire re evaluation of the concept of the gene.
The problem is that when the pieces of DNA are transcribed and then spliced together to make a master RNA copy with which to make proteins, this RNA can come from overlapping and totally unconnected pieces of DNA that used to be called discrete genes. This final spliced and edited piece of RNA then makes the protein. Perhaps, some scientists now say, this piece of finally spliced RNA should be considered the real “gene”.
The following will attempt to describe more of the findings. But, perhaps the most important finding from these 30 papers is how little we know about how all of this might work. This post and the next will attempt to make sense of it.
New Landscape of Genetic Regulation
The new landscape is drastically different and more complex.
A gene’s regulation is influenced by multiple stretches of regulatory DNA located both near and far from the gene itself and by strands of RNA not translated into proteins, so-called noncoding RNA (see previous post for a lengthy list of some of the newly discovered non coding RNAs).
 The ENCODE project specifically focused on the unknown 98% of the DNA and found 80% of the DNA was biologically active. There is much debate right now among geneticists about what “biologically active” implies. Does this mean all of the active DNA is relevant to regulation? Or could it mean that active DNA sequences can still be “junk”, meaning not doing anything important, remnants of previously useful steps from evolution.
The ENCODE project specifically focused on the unknown 98% of the DNA and found 80% of the DNA was biologically active. There is much debate right now among geneticists about what “biologically active” implies. Does this mean all of the active DNA is relevant to regulation? Or could it mean that active DNA sequences can still be “junk”, meaning not doing anything important, remnants of previously useful steps from evolution.
There is a major controversy now between those scientists who believe that most of the 80% is important and those who think that most is not. This question will be addressed further in the next post.
Certainly, however, it can be concluded that much more of the “junk” is important, more than anyone previously thought.
At Least Twenty Percent is Coding
ENCODE determined that 20% of the supposed junk DNA is involved in coding for 4 million regulatory switches. These are places in the DNA where proteins, or other molecules, stick to the DNA and cause an effect, possibly from the 70,000 promoters (affecting nearby genes) or the more than 400,000 enhancers (affecting far away genes) that have been discovered.
This 20% of definitely relevant switches is a very large number when you consider that it is only a portion of the regulatory elements involved. Twenty percent of definite switches means that regulation occupies 20 to 30 times the amount of the DNA as all of the “genes” themselves.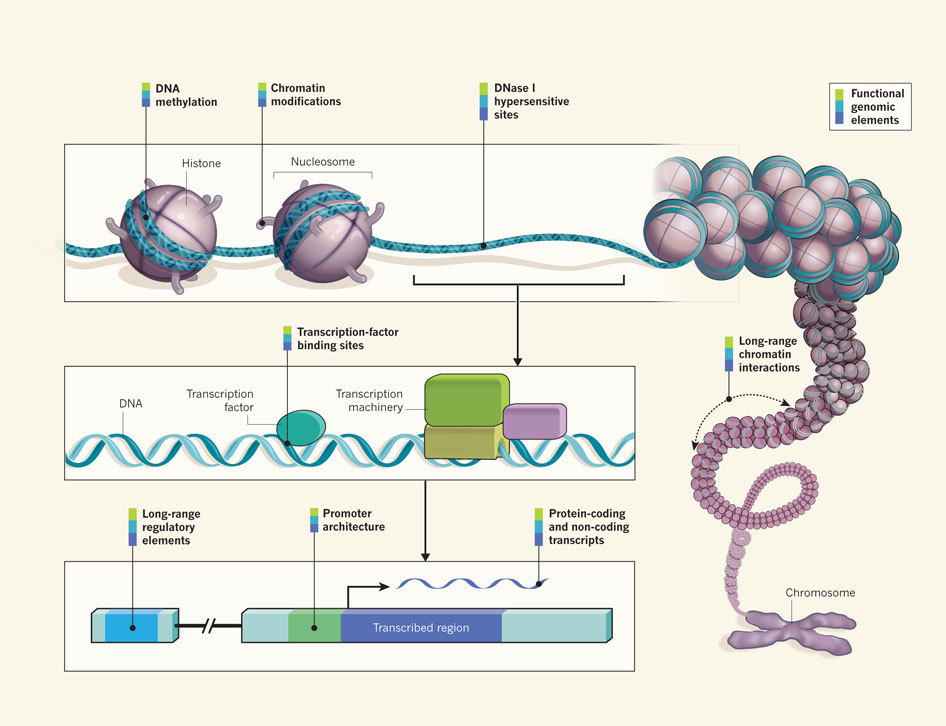
This figure has to be put into perspective. Previously it was believed that most of the regulation occurs with specific proteins mentioned above called transcription factors. Now, in addition to these protein factors, there is a regulatory region of DNA 20 times larger than all of the protein coding sections.
Another finding is one million different switches that can act in multiple different places. This greatly complicates the regulation. Just as transcripts are made from multiple different exons from different and overlapping “genes”, here a huge number of switches affect multiple different genes, not just one. The details of these switches will be described in the next post.
 Of the huge amount of new territory that was analyzed in the study, 18,400 regions were found that make definite RNA molecules of importance, 8800 small RNA molecules and 9600 long RNA molecules over 200 bases. These 18,400 regions that make critical definite RNA molecules are now called “RNA genes”. Please see the previous post for a sampling of newly discovered RNAs before ENCODE.
Of the huge amount of new territory that was analyzed in the study, 18,400 regions were found that make definite RNA molecules of importance, 8800 small RNA molecules and 9600 long RNA molecules over 200 bases. These 18,400 regions that make critical definite RNA molecules are now called “RNA genes”. Please see the previous post for a sampling of newly discovered RNAs before ENCODE.
To summarize: Now we have 80% of the DNA definitely biologically active, at least 20% definitely involved in regulation (perhaps much more), 4 million switches, 20,000 genes that make proteins, and 18,400 “RNA genes” that make definite functional RNA involved in regulation.
What Types of Cells?
 It is also important to note that ENCODE utilized 140 different types of cells for the analysis. These were cells that could be easily grown in a laboratory culture, which is a severe limitation. Neurons, for example, cannot be grown in culture and therefore were not directly studied. Also, each cell came from a different person. In future, it would be important to see if individuals regulate their cells differently from other people, and to see how one person regulates many different types of cells. In other studies the differences between cells were greater than the differences between people. Also, some of ENCODE’s cells are unnatural and are lines of cancer cells. One criticism of the study thus far is that the choice of the cells that were studied has been arbitrary.
It is also important to note that ENCODE utilized 140 different types of cells for the analysis. These were cells that could be easily grown in a laboratory culture, which is a severe limitation. Neurons, for example, cannot be grown in culture and therefore were not directly studied. Also, each cell came from a different person. In future, it would be important to see if individuals regulate their cells differently from other people, and to see how one person regulates many different types of cells. In other studies the differences between cells were greater than the differences between people. Also, some of ENCODE’s cells are unnatural and are lines of cancer cells. One criticism of the study thus far is that the choice of the cells that were studied has been arbitrary.
A striking early finding is that each type of cell had different regulation systems, which is logical but creates even more complexity because no one predicted the extent of this regulation. The kidney cell and the brain cell use the same DNA but regulate it in very different ways. The cells chosen for ENCODE are not the common human cell types because many don’t easily grow in culture. It is, therefore, not possible to extrapolate exact details about what will occur in the other types of human cells, but the principles are clear from the study.
Evolution and ENCODE
 This is such a vast rewrite of genetics in such a short time, the implications for evolution are not yet totally clear. Certainly, if most of the “biologically active” 80% are important it will have major implications for current evolution theory. But, this is not at all certain and is being actively debated. The next post will raise more issues about this biologically active 80%.
This is such a vast rewrite of genetics in such a short time, the implications for evolution are not yet totally clear. Certainly, if most of the “biologically active” 80% are important it will have major implications for current evolution theory. But, this is not at all certain and is being actively debated. The next post will raise more issues about this biologically active 80%.
The old evolutionary theory of disease assumed that mutations in genes, along with environmental factors, caused the abnormalities of diseases. But, ten years after the genome project, very few new diseases were found in the genes. And other types of gene research, such as the GWAs or genome wide association studies where genetic variants in many people are associated with a disease or trait, seemed to point to regions of the junk DNA as relevant to these diseases.
With the new ENCODE data it is clear that in different types of cells, there are different very complex regulatory networks in up to 80% of non coding, non “gene” DNA. It is now thought that most of the mutations of common diseases will be in the regulatory regions.
Mind and Cell
 Mental events stimulate immediate action in the molecular machinery of the individual neurons, rapidly manufacturing and transporting complex molecules to all regions of the cell. Some of the regulation is now identified to be among 4 million possible switches, each interacting with multiple different sites, and using pieces of overlapping and disconnected genes.
Mental events stimulate immediate action in the molecular machinery of the individual neurons, rapidly manufacturing and transporting complex molecules to all regions of the cell. Some of the regulation is now identified to be among 4 million possible switches, each interacting with multiple different sites, and using pieces of overlapping and disconnected genes.
ENCODE is expanding the scope of the regulation in a neuron’s genetic machinery, making it even more difficult to determine how mind instantly triggers these changes.
The next post will continue with more very interesting details from ENCODE.