 To pass on the code of life to the next cell, DNA copies itself. This process is called replication. Much is made of the mutations, or errors in DNA replication. Evolutionary theory relies in part on these mutations to explain the development of the dramatic diversity of nature; however, what is most dramatic about DNA is not its errors but its accuracy. Many levels of proofreading and error correction ensure near-perfect fidelity in replication.
To pass on the code of life to the next cell, DNA copies itself. This process is called replication. Much is made of the mutations, or errors in DNA replication. Evolutionary theory relies in part on these mutations to explain the development of the dramatic diversity of nature; however, what is most dramatic about DNA is not its errors but its accuracy. Many levels of proofreading and error correction ensure near-perfect fidelity in replication.
Current theory suggests DNA somehow directs the entire replication process, perhaps through RNA messages. But, since there is editing and error correction involving the DNA itself, it is hard to imagine exactly how this is done. Regulation for these processes is massively complex; currently, there is no obvious source of direction.
DNA Errors and Proofreading
 During replication, nucleotides, which compose DNA, are copied. When E coli makes a copy of its DNA, it makes approximately one mistake for every billion new nucleotides. It can copy about 2000 letters per second, finishing the entire replication process in less than an hour. Compared to human engineering, this error rate is amazingly low. E coli makes so few errors because DNA is proofread in multiple ways.
During replication, nucleotides, which compose DNA, are copied. When E coli makes a copy of its DNA, it makes approximately one mistake for every billion new nucleotides. It can copy about 2000 letters per second, finishing the entire replication process in less than an hour. Compared to human engineering, this error rate is amazingly low. E coli makes so few errors because DNA is proofread in multiple ways.
An enzyme, DNA polymerase, moves along the DNA strands to start copying the code from each strand of DNA. This process has an error rate of about one in 100,000: rather high. When an error occurs, though, DNA polymerase senses the irregularity as a distortion of the new DNA’s structure, and stops what it is doing. How a protein can sense this is not clear.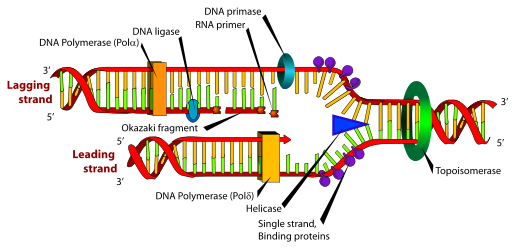
Other molecules then come to fix the mistake, removing the mistaken nucleotide base and replacing it with the correct one. After correction, the polymerase proceeds. This correction mechanism increases the accuracy 100 to 1000 times.
A Second Round of Proofreading
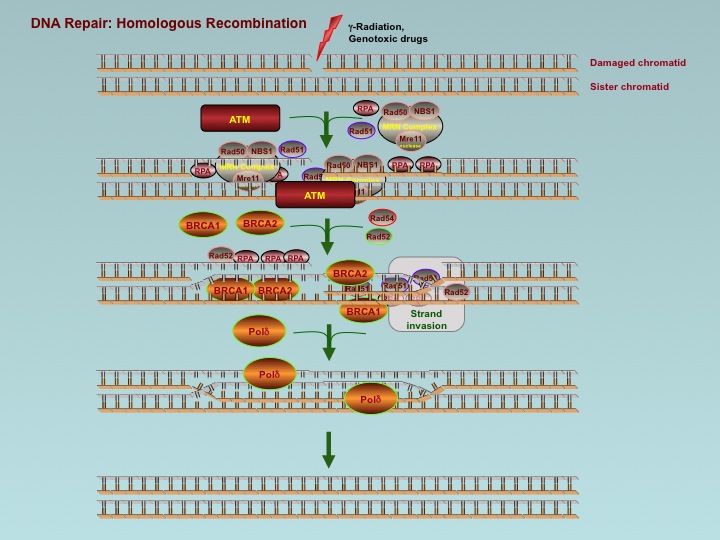
There are still some errors, however, that escape the previous mechanism. For those, three other complex proteins go over the newly copied DNA sequence. The first protein, called MutS (for mutator), senses a distortion in the helix shape of the new DNA and binds to the region with the mistaken nucleotides. The second protein, MutL, senses that its brother S is attached and brings a third protein over and attaches the two. The third molecule actually cuts the mistake on both sides. The three proteins then tag the incorrect section with a methyl group.
Meanwhile, another partial strand of DNA is being created for the region in question, and another set of proteins cut out the exact amount of DNA needed to fill the gap. With both the mistaken piece and newly minted correct piece present, yet another protein determines which is the correct one by way of the methyl tag. That is, the correct one does not have the methyl tag on it. This new, correct section is then brought over and added to the original DNA strand.
This second proofreading is itself 99% efficient and increases the overall accuracy of replication by another 100 times. Below is a diagram showing the complexity of one of the proofreading mechanisms (one out of many different editing mechanisms.)
Multiple Sensors
There are multiple places where a protein “senses” what needs to be done. The computer-like sensing of the original mistake, cannot be directed by the original DNA. Clearly, there are other sources of decision-making in a cell.
While DNA’s “quality control” is extremely complex in E.Coli, the same process is even more complex in the human cell. Human cells contain many different polymerases and many other enzymes to cut and mend mistakes. There are even different Mut-type systems that, along with other proofreading, render human DNA replication incredibly accurate.
Very recent research has shown some of the complex mechanisms of the MutL family of mutation correction molecules. It shows that an energy molecule ATP stimulates the process whereby MutL cuts the DNA around the error. There are two grooves in the MutL molecule, one for ATP and one for the DNA strand. When ATP binds to MutL it changes the protein’s shape which allows the cutting to occur. In humans when MutL is not functioning properly it is know to cause cancer.
Many Mutation Causes
 The meaning of the word “mutation” has evolved to refer to any changes in the DNA sequence in a cell’s genome. There are many different causes of mutations in the extremely long and complex DNA code. In each cell there is 2 yards of double stranded DNA with segments tightly wrapped around many small balls of protein called nucleosomes (histones are a well known molecule that is a part of a nucleosome). Mutations that are incorporated into all the cells in a human beings body cause major diseases such as Huntington’s disease, Fragile X, Sickle-cell disease, and many others. Mutations that arise as cells copy themselves in the blood, skin, immune system, gut, and to some extent brain, can lead to changes that lead to cancer. In cancer the cells with the new mutations continue growing new cells.
The meaning of the word “mutation” has evolved to refer to any changes in the DNA sequence in a cell’s genome. There are many different causes of mutations in the extremely long and complex DNA code. In each cell there is 2 yards of double stranded DNA with segments tightly wrapped around many small balls of protein called nucleosomes (histones are a well known molecule that is a part of a nucleosome). Mutations that are incorporated into all the cells in a human beings body cause major diseases such as Huntington’s disease, Fragile X, Sickle-cell disease, and many others. Mutations that arise as cells copy themselves in the blood, skin, immune system, gut, and to some extent brain, can lead to changes that lead to cancer. In cancer the cells with the new mutations continue growing new cells.
Cigarettes, car exhaust, Ultraviolet light, radiation, viruses, transposons (jumping genes), oxidants and a wide variety of toxic carcinogens all damage the DNA in various ways. Some cells, when in a difficult situation and needing a change, seem to induce increased mutations in a process called hypermutation. Some of the  mutations are simply mechanical errors such as small bits of RNA being included in the DNA. Some are DNA duplications, where a sequence of code is copied one or more times and then included back in the DNA. Other mutations are incorrect copying of a letter in the sequence, where one letter is exchanged for another. Copying DNA involves many different steps including deliberate splicing of smaller pieces together and so there are many different places where mutations can occur in the process.
mutations are simply mechanical errors such as small bits of RNA being included in the DNA. Some are DNA duplications, where a sequence of code is copied one or more times and then included back in the DNA. Other mutations are incorrect copying of a letter in the sequence, where one letter is exchanged for another. Copying DNA involves many different steps including deliberate splicing of smaller pieces together and so there are many different places where mutations can occur in the process.
Given the amount of varied mutations what is truly remarkable is that such accurate proofreading can occur.
And Many Editing Solutions
There are many overlapping editing processes for the multiple steps in the complex task of copying DNA. One of the steps in replication of DNA is opening up the very tightly bound, protein balls protecting the DNA strand. Another step involves the polymerase enzyme copying both strands. This step is confounded by the fact that the two strands are in opposite directions and the copying goes in the same direction on both. The strand that is copied backwards is broken into many different pieces which are then spliced together, another source of errors.
Many of these mechanisms are just being discovered.
The most common mistake, the inclusion of a small bit of RNA in the DNA a million times in each cell division, was recently noted to have a special enzyme for this repair. RNA’s pieces are very similar to DNA but less stable and therefore need to be corrected.
Another process was recently discovered utilizing the very important protein ubiquitin, usually the workhorse that tags unneeded molecules for destruction. In this new process ubiquitin tags a DNA break site for other molecules to fix. One step that was recently discovered in this process is the mechanism that stops the repair process when it is done. While initiation of this process is critical, equally important is stopping the process so that it doesn’t keep operating on areas that do not need repairs. A complex mechanism is described where one subunit pushes off another cancelling the repair process.
In another process, the enzyme DNA ligase has been shown to encircle DNA, a technique in the critical step of rejoining DNA after repair is done. There are specific grooves in this protein which bind on both the upstream and downstream ends of the part of the DNA that is broken. Ligase has a special groove for ATP molecules to power the process and a section that bonds the two parts of the DNA back together.
Yet another mechanism of repair was recently found when the DNA strand breaks. It involves two proteins working together. In bacteria the molecule RecA attaches to one of the broken ends of the DNA and forms a filament. This filament searches for the other disconnected DNA strand. It attaches to all pieces of DNA that happen to be nearby and studies the strands to determine the one that has the exact sequence that is needed to replace the broken DNA. When a correct sequence is found, the molecule binds the two ends and waits for the second molecule to help attach them.
A very recent study found another complex sensor of abnormal DNA. Several molecules of two specific proteins, called UvrA and UvrB, surround the DNA and form a tunnel between them for the DNA. The sensor complex then “stresses” the DNA by clamping down on the strand. If the entire system of proteins surrounding the DNA can change shape to a closed form then the DNA is normal. When the system remains in an open state the DNA is abnormal and an excision repair process is stimulated.
Cellular Self Engineering
While mutations help determine evolutionary variety, we still don’t know how these very elaborate and multi-layered quality controls came about and how they are directed. Is it possible for DNA to directed its own editing? Somehow, these processes know which are appropriate DNA sequences and which are not.
Mutations help produce variety in evolution. If a cell can in any way control and direct the mutations through these elaborate control and editing functions, then survival can be improved. Previous posts have mentioned microbes’ sentient ability to communicate, make decisions, and mount defenses. It has been suggested that this editing is a form of self directed engineering by the cell (see resources, Evolution: A view from the 21st Century.)
Is the self directed, complex proofreading of DNA replication another form of cellular sentience? If it is how has it influenced evolution?