 How does the mind work in an instant through billions of neurons, trillions of synapses, and millions of interacting switches deep inside cells? The regulation of molecular genetic processes has been shown to be vastly more complex than previously thought.
How does the mind work in an instant through billions of neurons, trillions of synapses, and millions of interacting switches deep inside cells? The regulation of molecular genetic processes has been shown to be vastly more complex than previously thought.
What do these new findings imply about the mechanism of brain evolution?
1.5% of the DNA make proteins, the so called 20,000 “genes”.
80% of all the DNA is biologically active, that is, it actually makes RNA.
At least 20% of the biologically active DNA is definitely involved in regulation. This amount is at least 15 times greater than the amount of DNA making proteins.
 At least 4 million switches are active, often operating in multiple different places at once.
At least 4 million switches are active, often operating in multiple different places at once.
18,000 places in the DNA where functional RNA is made, the new “RNA genes.” 8 million different particles interact and regulate these RNAs.
To make a protein pieces of DNA are taken from multiple regions, not just one “gene.” RNA then edits these pieces into a working template.
Digesting the impact of the 30 ENCODE (Encyclopedia of DNA Elements) studies will be difficult both for scientists and for the general population. In order to understand how mental events can instantly stimulate genetic machinery the details of the new genetic landscape will need to be incorporated into neurochemistry theory.
 It is impossible to comprehend how the fetal brain can create and integrate 250,000 neurons each second during the last month of pregnancy. It is also impossible to tabulate 100 billion neurons each connected with thousands of other neurons, the synapses changing each day.
It is impossible to comprehend how the fetal brain can create and integrate 250,000 neurons each second during the last month of pregnancy. It is also impossible to tabulate 100 billion neurons each connected with thousands of other neurons, the synapses changing each day.
Understanding how four million interacting DNA switches and 8 million particles regulating RNA operate is equally difficult. But, the regulation of genetic material is really much more complex than just the millions of switches.
A new study showed that each type of neuron, and each region of the brain, have different patterns of transcription further complicating this picture.
Here we will elaborate several more details from ENCODE.
More DNA Elements
While somewhat confusing, these are part of the new language of the cell and therefore are being mentioned.
Transcription Factors – Switches Made of Proteins
Proteins, called transcription factors, act as switches. They bind to DNA and determine whether RNA is made or not. These can be promoters, which activate DNA, or repressors, which block action. Other proteins that act on DNA include co-activators, chromatin remodelers, histone enzymes, kinases that transfer energy phosphate molecules, and methylases that add a methyl group.
ENCODE found 4 million DNA sites, 8% of the DNA, where these factors operate. But, there were 45 million different events where transcription factors occupied DNA. This real number is probably much greater since only certain cells and places within cells have been studied. It is not clear how many of these are active or evolutionarily old switches that are not used now.
These individual switches almost all interact with many genes, and most genes have multiple switches acting upon it at the same time.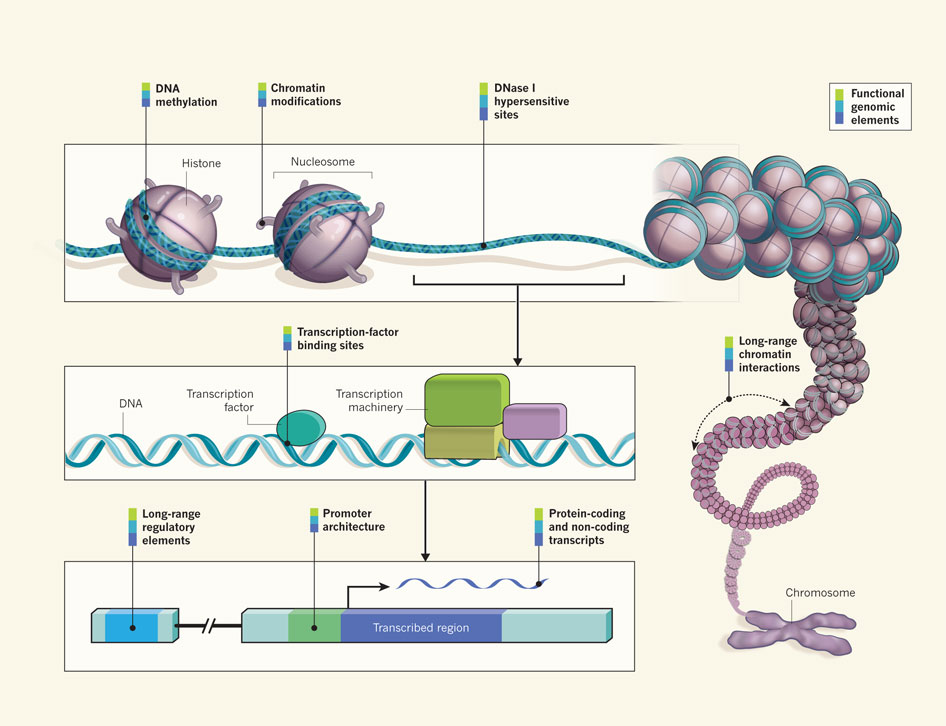
DNA methylation
These markings on DNA are an important part of epigenetics, that is actions other than DNA that affect heredity. Attaching methyl groups to DNA sections blocks DNA making proteins. Methylation of specific genes is crucial in determining long-range effects of many experiences in life including starvation, stress, and abuse.
Open Chromatin
DNA is tightly bound, wound and looped by multiple large proteins, which in total are called chromatin. Histones are one important large molecule involved in winding and unwinding DNA. Only when DNA is open can it be acted upon. Mechanisms that open the tightly bound DNA allow the DNA to be influenced by the regulatory particles. Somehow, the cell knows exactly where it wants to unfold these loops to find the perfect DNA for the occasion.
Modified histones
There are many modifications that can alter histones, which then change the ability to use DNA. Sixty different modifications have now been identified, and 13 of these have been described in the ENCODE research.
RNA Binding
This is a place where regulatory proteins attach to RNA. ENCODE identified 8 million distinct particles.
DHS (DNase Hypersensitivity Sites)
Hypersensitivity sites are those areas on the DNA where there is no interference from histone molecules. ENCODE identified 200,000 sites per cell type. It also identified three million places where transcription factors bind to these 200,000 open sites. This means that 42% of the DNA that codes for protein is accessible to switching. These sites are very different in different types of cells. ENCODE has found 500,000 enhancers to go with these sites out of 3 million identified but not yet detailed.
Most of these sites are very highly regulated. Each site has dozens to hundreds of co activators. 88% of DHS are active during development of the fetus, and respond with variations based upon exposures.
Additional ENCODE factors
There are additional confounding complications that ENCODE identified:
1. There are many very distant regulators. It is important to realize that most of the study of DNA is in two dimensions and yet in reality it is looped and perhaps some regions that seem distant are closer in three dimensions.
2. RNA pieces are spliced together while they are being transcribed. Surprisingly, microRNA, which are important regulators, are also spliced together. Splicing creates an entirely different level of complexity. Where is the direction and regulation of this splicing? It is especially perplexing why small RNAs would be spliced together since these seem to be critical regulatory factors to begin with. What is regulating the regulators?
3. Another confounding development is that transcription factors are now shown to bind at different levels. Therefore, it is not a simple “on” and “off” switch but a dimmer switch at different levels of intensity. Also, they operate in different ways in different circumstances with multiple levels of feedback loops determining their actions in the circuits they influence.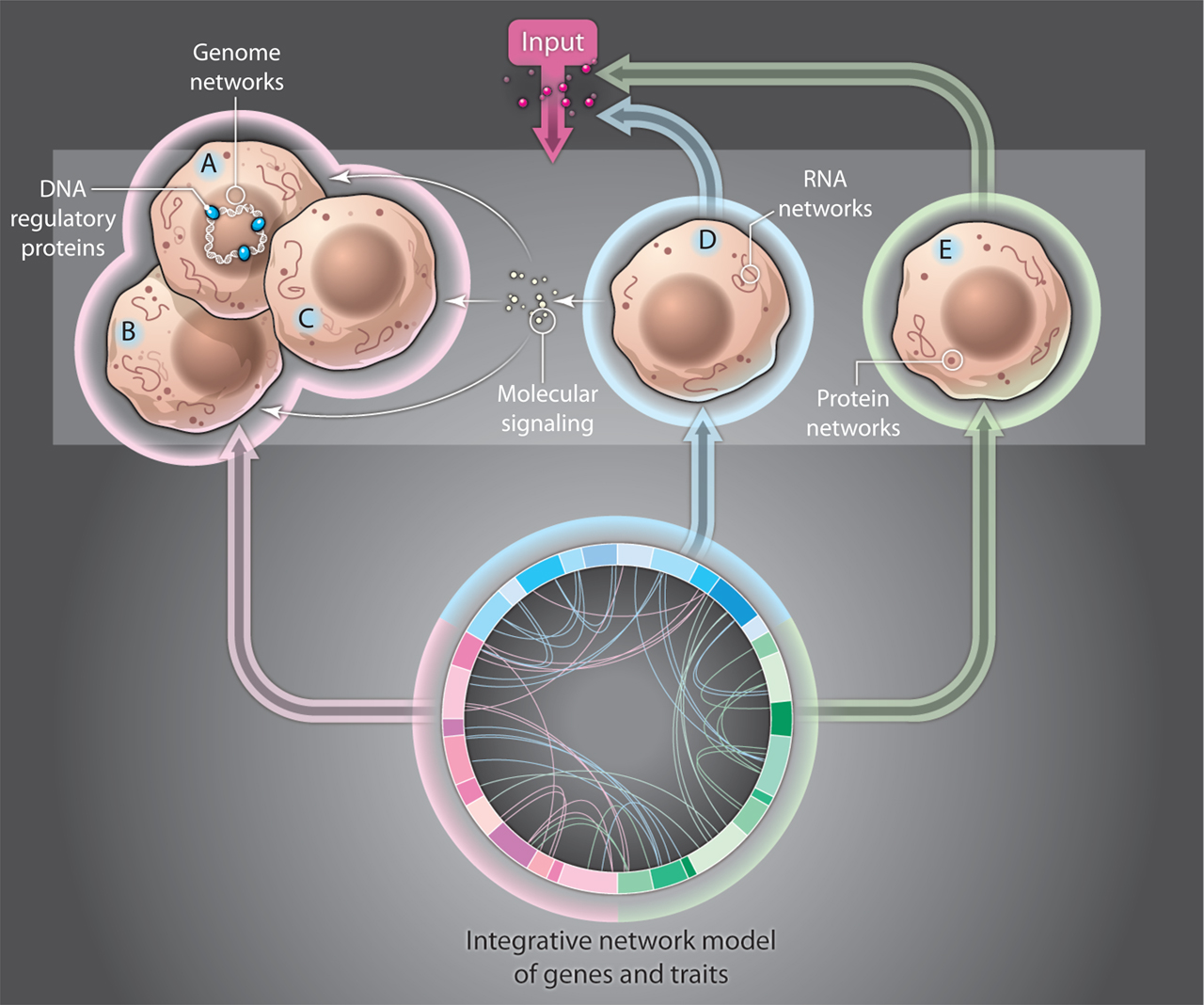
4. It is not at all clear how the vast number of small RNAs know how to get to the specific addresses in the cell where they operate.
5. The same regulatory region can play very different roles in different types of cells, and at different times
Evolution
 Previously, individual mutations in protein coding regions genes (see previous post) were expected to be the major driver of evolution. But, protein coding DNA tends to be very stable over long stretches of evolution. There are many regions of DNA that are similar in humans and mice, and some even in microbes. From individual mutations, the rate of change of the DNA that codes for proteins has been much too slow to explain the changes in evolution. Also, searches for causes of diseases in the protein coding genes have just not been productive.
Previously, individual mutations in protein coding regions genes (see previous post) were expected to be the major driver of evolution. But, protein coding DNA tends to be very stable over long stretches of evolution. There are many regions of DNA that are similar in humans and mice, and some even in microbes. From individual mutations, the rate of change of the DNA that codes for proteins has been much too slow to explain the changes in evolution. Also, searches for causes of diseases in the protein coding genes have just not been productive.
But, the new huge regulatory regions are a different story. There is much greater turnover and innovation in the regulatory regions. In a word, evolution is occurring much faster in the vast regulatory regions. And the huge increase in regulation in humans might explain much of the differences  from other creatures.
from other creatures.
A critical unanswered question is whether the 80% biologically active section of the DNA is junk or functioning RNA. ENCODE clearly showed that 80% is active enough to make pieces of RNA, and that 20% of this RNA is definitely purposeful, that is, engaged in specific regulation. But, possibly much more than 20% is very relevant. The greater the percentage, the less random and messy the evolution process.
Junk DNA or a Jungle of DNA?
 The chief scientist of ENCODE recently said that the word “junk” should be put to rest, and the new word might be “jungle”.
The chief scientist of ENCODE recently said that the word “junk” should be put to rest, and the new word might be “jungle”.
Some of the junk areas are old genes that no longer work, called “pseudogenes”.
Another type of junk has been mobile elements. These pieces of DNA, called transposons, retrotransposons, or “jumping genes,” are virus-like particles that are able to use enzymes to cut themselves out, move and replace themselves into the DNA strand. Some of these jumping genes stay where they are but use enzymes to make copies, which travel and place themselves into another region of DNA. Some make many copies that greatly enlarge the amount of DNA in a creature. Some of these seem to be random, but some might have a specific function.
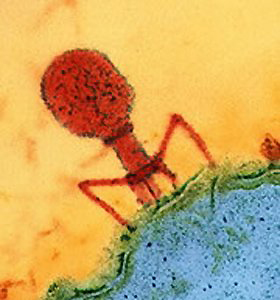 The action of these DNA pieces is similar to “horizontal gene transfer” (discussed in posts on microbes and viruses) that is extremely common in microbes, sometimes using phage viruses, or phage virus like particles to transfer DNA between cells. It is well known that microbes use these virus like DNA transfers to share innovations, such as antibiotic resistance.
The action of these DNA pieces is similar to “horizontal gene transfer” (discussed in posts on microbes and viruses) that is extremely common in microbes, sometimes using phage viruses, or phage virus like particles to transfer DNA between cells. It is well known that microbes use these virus like DNA transfers to share innovations, such as antibiotic resistance.
Increasingly, horizontal transfer is being viewed as significant for larger multi-celled creatures including humans. The transfer of genetic material is observed between viruses or microbe cells, and human cells. There is also increasing evidence that microbes in the human’s body greatly influence the host by producing factors that affect all human organs including the brain.
There are many mobile genes in the human DNA. What is not known is how many of these are junk versus usable new information. It is also not known how many are involved in the cell’s self-editing.
An important new finding has been that evolutionary change in proteins doesn’t usually occur with simple one amino acid point mutations, but rather by whole new sections, called domains. (Simple single mutations mostly are destructive.) Domains are a sequence of amino acid, like an branch of the protein, that can bind to another molecule, changing the function of protein. Jumping genes, or other DNA transfers, can take the entire genetic section of a domain and move it, add it or subtract it, changing the entire function of the protein. A similar mechanism is used by microbes to transfer the ability to fight an antibiotic to other cells. For protein re-design and evolution, mobile DNA elements, could be a prime mechanism.
Cellular self-editing, cellular decision-making, and mobile DNA, have major implications for evolution theory. ENCODE shows that most of these are active, now it has to be determined if they are relevant.
Thought Activates a Neuron
 When a thought affects a neuron, vast genetic regulation activity occurs in an instant. Millions of interacting switches in billions of neurons with trillions of synapses respond instantaneously. DNA and RNA manufacture microtubules, which build the neuron’s highways, carrying building materials and sacs of neurotransmitters to the budding dendrite, merging the sacs of neurotransmitters with the membrane to release the neurotransmitter. Countless synchronized ion channels are opening and closing.
When a thought affects a neuron, vast genetic regulation activity occurs in an instant. Millions of interacting switches in billions of neurons with trillions of synapses respond instantaneously. DNA and RNA manufacture microtubules, which build the neuron’s highways, carrying building materials and sacs of neurotransmitters to the budding dendrite, merging the sacs of neurotransmitters with the membrane to release the neurotransmitter. Countless synchronized ion channels are opening and closing.
All of this involves the regulatory activity described by ENCODE. For this level of cellular activity to exist, cells must indeed be very intelligent.