 Which comes first the chicken or the egg? With proteins and RNA regulating the function of DNA and DNA producing RNA which then produces proteins, which comes first—DNA, RNA, or proteins? At first, the way DNA makes proteins seemed almost clear and simple—DNA makes RNA, which makes proteins in one direction.
Which comes first the chicken or the egg? With proteins and RNA regulating the function of DNA and DNA producing RNA which then produces proteins, which comes first—DNA, RNA, or proteins? At first, the way DNA makes proteins seemed almost clear and simple—DNA makes RNA, which makes proteins in one direction.
But, now multiple vast networks of regulation have been found that include more than forty different types of tags placed on both DNA and the protective histone proteins that allow access to DNA strands. All of these tags alter genetic function and are based on the actions of protein enzymes.
 Another type of regulation involves the shape of the DNA both in small loops and large placement inside of highly structured nucleus scaffolds with dramatic influence on genetic functions. The very complex editing of messenger RNA can make up to 500 different proteins from a single strand of DNA—what used to be called a gene, but now is hard to define. Genes can overlap and triggers of regulation can happen in the gene or in the regulatory strand.
Another type of regulation involves the shape of the DNA both in small loops and large placement inside of highly structured nucleus scaffolds with dramatic influence on genetic functions. The very complex editing of messenger RNA can make up to 500 different proteins from a single strand of DNA—what used to be called a gene, but now is hard to define. Genes can overlap and triggers of regulation can happen in the gene or in the regulatory strand.
Where is the regulation for this?
 What is equally amazing is that mental events—thought—almost instantly triggers large networks of many genes at the same time. How does this happen? See previous posts for some descriptions of this (The life of a thought in the brain, More complexity in genetic Regulation)
What is equally amazing is that mental events—thought—almost instantly triggers large networks of many genes at the same time. How does this happen? See previous posts for some descriptions of this (The life of a thought in the brain, More complexity in genetic Regulation)
Now entirely new sets of factors have appeared that are based on physical properties of the coding of RNA and the liquid phase transitions that separate active and inactive sections of the chromosomes. The first unusual new physical properties occur when there are repeated patterns of code. The regions of regulatory code where there are large amounts of repeats have been correlated with multiple different degenerative diseases such as Huntington’s and ALS. The second is the way that active and inactive sections of the DNA are in different regions of the nucleus.
This post will address this new research that adds to new complexity with DNA regulation.
First a summary list of the fantastic complexity of regulation of DNA and then the very new research on how clusters of RNA strands have unique physical properties that are somehow related to this regulation.
Summary List of Factors that Regulate DNA
It is not known how many overlapping layers of regulation there are there are:
Some include:

From Smallbot Histones protect DNA and must be opened to be used. There are now forty different tags that determine whether histones can be opened or not to utilize DNA.
- There are many newly discovered tags on DNA itself that alter function.
- Thousands of proteins called promoters and enhancers start and stop DNA function.
- Promoters work in many different places for each gene and in different places for each type of human cell. Also, for even more complexity, multiple promoters combine into larger machines, some touching the DNA and some only combining with the other promoters. Multiple structures exist for each gene in different places.
- There are multiple different places where transcription starts for each “gene”, again undermining the notion of the simple “gene.”
- Thousands of large and small RNAs interact in multiple ways to regulate what pieces of DNA are used.
- Many different mechanisms are used to repair DNA errors without obvious direction.
- Eight million factors affect the RNA particles that are made from at least 20% of all DNA (maybe up to 50%). Messenger RNA somehow determines multiple different edits from the same pieces of DNA.
- Pieces of DNA and RNA are taken from multiple places, strands are cut out and others sewn together without clear direction.
- In some DNA there are two superimposed codes at once in one section of DNA. These are related to new messenger RNA folding, and multi-use codons called “duons.” The first purpose is to transcribe DNA code to RNA code to make proteins. The second purpose is to bind regulatory factors, as the regulatory regions do to trigger other genes.
 Transcription factors bind inside at least 13% of “genes” themselves, not just the regulatory regions nearby. This finding was truly shocking since the DNA in the gene, then, must code for two meanings at once for two entirely different purposes.
Transcription factors bind inside at least 13% of “genes” themselves, not just the regulatory regions nearby. This finding was truly shocking since the DNA in the gene, then, must code for two meanings at once for two entirely different purposes.- RNA splicing, can make up to 500 different proteins from the same pieces of DNA (see post). The RNA cuts out sections and sews others together. Recently, it has been demonstrated that the pieces that are sewn together by the messenger RNA may come from multiple different areas that used to be called individual genes.
- Four million different switches are active, often operating in multiple places at once; 18,000 places where active RNA is made; and 8 million different particles interacting to regulate these RNAs.
- The entire concept of “gene” has to be reconsidered because messenger RNA takes pieces from different regions to make one RNA edit for a protein.
- 3D shapes of RNA have great importance in the process. Hairpin loops where regions of RNA have complementary sections of RNA code. This loop folding forms a secondary structure that appears to have significance in translation of RNA code into proteins. The studies showed that even closely related individuals could have different folding structures. 1900 different mutations were identified that affect these RNA structures
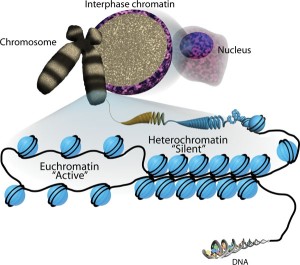
- The 3D shape of the DNA chromosome is correlated with the activity of the genes inside.
 The links exist at multiple levels.
The links exist at multiple levels.- Loops of chromatin make space for enhancers to land. A larger relationship exists for specific places in the nucleus to impact on particular active DNA sites.
- Special protein structures alter polymerase activity.
- Chromosomes with fewer active genes are placed at the edge of the nucleus while the active ones are near the center.
- Less active sections are placed near the nuclear lamina, which is close to membrane.
- The location can suppress the genetic machinery.
- If the chromatin is opened but not used, the entire section is moved to a different location.
- The exact location of the gene in space in the nucleus influences its activity.
- To fit 2 yards of DNA into a tiny nucleus is a monumental engineering feat. DNA is highly compacted yet has to be instantly available to rapidly make proteins in neurons with a momentary change of thought.
New Development: Clusters of RNA form Special Droplets
 A normal section of DNA can accumulate repeats of small sections that don’t code for proteins and somehow cause brain damage in ALS and Huntington’s. It appears to be related to the number of repeats and there has not, until now, been any way to understand why this threshold existed. Now it has been learned that the repeats code RNA strands with similar repeats have unique physical properties.
A normal section of DNA can accumulate repeats of small sections that don’t code for proteins and somehow cause brain damage in ALS and Huntington’s. It appears to be related to the number of repeats and there has not, until now, been any way to understand why this threshold existed. Now it has been learned that the repeats code RNA strands with similar repeats have unique physical properties.
These RNA repeats can form clusters in the form of a ball. What is highly unusual is that these balls of repeated RNA change their physical state into a gel like structure. This unique structure is made up of the RNA plus some proteins. Its form is like a liquid that is very compacted. This has been likened to morning dew where water forms droplets. It goes by a complex title—liquid-liquid phase separation or LLPS. This cluster is like an organelle in some ways but has no membrane and is created by chemical bonds between the RNA repeats and the proteins. Alterations in the proteins for specific brain diseases also alter the condensed liquid into a gel.
When the specific repeats for these brain diseases were evaluated, they were found to form this gel in the form of a ball. But, what was seen is that this only occurs when enough of the repeats were present, which is important because Huntington’s disease and ALS only occur with over a specific number of repeats.
 Other research showed some of the repeats were altered and they didn’t form the gel clusters. They stayed irregular. Adding molecules also disrupted the entire gel process, which showed that the chemical interactions between the RNA base pairs is the basis of the cluster. In this case, they formed clusters that looked like the gel droplets, but were in fact solid without the special phase effect.
Other research showed some of the repeats were altered and they didn’t form the gel clusters. They stayed irregular. Adding molecules also disrupted the entire gel process, which showed that the chemical interactions between the RNA base pairs is the basis of the cluster. In this case, they formed clusters that looked like the gel droplets, but were in fact solid without the special phase effect.
When the numbers reached the threshold, RNAs in the cell formed these clusters in the newly discovered small organelle in the nucleus called the nuclear speckle. Speckles don’t have membranes but are new discovered structures that are needed for messenger RNA to make proteins. When the base interactions were blocked they stayed in the speckle but didn’t stop its function. Specific strands form different kinds of clusters, some liquid and some not.
As with all areas of genetic regulation, the entire picture is not at all clear. While this new structure seems important, ALS might also have a unique form translating RNA into proteins as well. How the gel fits in needs further research.
In Huntington’s repeats occur in coding strands producing both repeating proteins and RNAs. It is not known if both are involved in the disease or just one. Most current research has been in the proteins, now RNA needs more work.
These clusters might be involved in another very small organelle without membranes—paraspeckles—related to long non coding RNA. The paraspeckle is not well understood but appears to be important in the process of DNA making RNA.
Another New Development: Liquid Property Separates Active and Inactive Chromatin
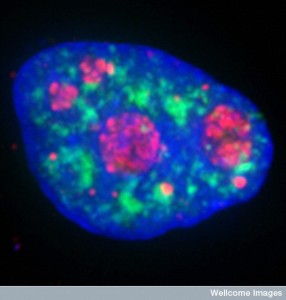 It now appears that in the nucleus different phase properties of liquids separates the active and inactive DNA chromosomes. These properties of liquids are well known in physics but only now shown to be relevant in biology. It is much the way quantum effects like photosynthesis and chemical reactions are now being discovered in biology.
It now appears that in the nucleus different phase properties of liquids separates the active and inactive DNA chromosomes. These properties of liquids are well known in physics but only now shown to be relevant in biology. It is much the way quantum effects like photosynthesis and chemical reactions are now being discovered in biology.
In the nucleus two kinds of liquids don’t mix—one with active genes making RNA and those that are inactive. These drops of liquid fuse together like oil would in water and seem to explain the organization of genes in the nucleus space. Very surprisingly, it seems to help regulate which genes are silent and which are active.
Large strands of DNA must be kept quiet for the correct gene to be triggered. This used to be thought to be based on histones and compaction techniques. It was thought that the tightly wound DNA would allow only the special enzymes to get at the particular DNA strands.
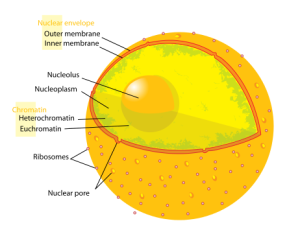 But, now it is observed that two different types of physical liquids that don’t mix in the nucleus have genes that are active and those that are not. The part that is not active becomes droplets that fuse together into larger quiet groups.
But, now it is observed that two different types of physical liquids that don’t mix in the nucleus have genes that are active and those that are not. The part that is not active becomes droplets that fuse together into larger quiet groups.
This also explains how large proteins can get access without having to deal with compaction and histones. Proteins go to one liquid environment or the other based on physical molecular characteristics such as the interactions of the atoms in the molecule related to temperature, concentration and pH. In diseased organs cellular structures are organized this way as well. These properties can change with age as well, where they become more solid and less liquid creating damage in the brain with clumps of proteins.
New Complexity in DNA Regulation
 Perhaps the most unusual aspect of all of this are the findings of how mental events trigger vast genetic changes. Human thought instantly alters large circuits of neurons (see post of neuroplasticity). This occurs through a wide variety of different neuroplasticity mechanisms by triggering specific gene networks and the creation of different complex machinery protein in each different brain region. Perceptions of loneliness are very significant triggers of gene networks in immune cells causing more inflammation—130 anti inflammatory genes have been shown to lack function and 80 pro inflammatory genes were very active.
Perhaps the most unusual aspect of all of this are the findings of how mental events trigger vast genetic changes. Human thought instantly alters large circuits of neurons (see post of neuroplasticity). This occurs through a wide variety of different neuroplasticity mechanisms by triggering specific gene networks and the creation of different complex machinery protein in each different brain region. Perceptions of loneliness are very significant triggers of gene networks in immune cells causing more inflammation—130 anti inflammatory genes have been shown to lack function and 80 pro inflammatory genes were very active.
Which of these layers of regulation allow mental activity to trigger so many genes in microseconds? Where is the direction for so many levels of action?
 Transcription factors bind inside at least 13% of “genes” themselves, not just the regulatory regions nearby. This finding was truly shocking since the DNA in the gene, then, must code for two meanings at once for two entirely different purposes.
Transcription factors bind inside at least 13% of “genes” themselves, not just the regulatory regions nearby. This finding was truly shocking since the DNA in the gene, then, must code for two meanings at once for two entirely different purposes.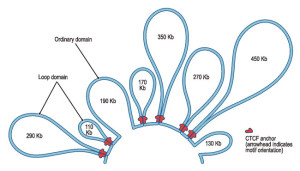 The links exist at multiple levels.
The links exist at multiple levels.