 Recent studies have described even more layers of codes and ways the genetic system is ordered in each cell. Two completely new superimposed codes have been described that greatly complicate genetic regulation—messenger RNA folding, and multi use codons called “duons.” In addition, this week the large international FANTOM project published 16 studies that demonstrate vast new complexity in the way DNA regions are triggered. In fact, more and more new studies reveal higher levels of genetic complexity.
Recent studies have described even more layers of codes and ways the genetic system is ordered in each cell. Two completely new superimposed codes have been described that greatly complicate genetic regulation—messenger RNA folding, and multi use codons called “duons.” In addition, this week the large international FANTOM project published 16 studies that demonstrate vast new complexity in the way DNA regions are triggered. In fact, more and more new studies reveal higher levels of genetic complexity.
Human perceptions alter networks of genes in cells throughout the brain, immune system and other organs, even though humans are six orders of magnitude larger than DNA genetic networks. While thought and perception immediately alter large brain circuits with many different neuroplasticity mechanisms at once, they do this by triggering gene networks deep inside all the cells in the circuit.
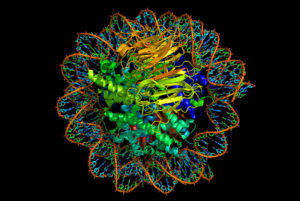
 When stimulating the use of DNA to manufacture new proteins, the protective protein spools, called histones, must unravel the DNA strands that are wrapped around the four large complex molecules. Markings on the histones by acetyl molecules form a code that determines whether a section of DNA will open or not. There are many other regulatory factors, including small and large RNA molecules that, also, influence whether DNA unravels from the histone.
When stimulating the use of DNA to manufacture new proteins, the protective protein spools, called histones, must unravel the DNA strands that are wrapped around the four large complex molecules. Markings on the histones by acetyl molecules form a code that determines whether a section of DNA will open or not. There are many other regulatory factors, including small and large RNA molecules that, also, influence whether DNA unravels from the histone.
When the DNA is unravelled and open, markings of methyl molecules attached to the DNA determine whether the specific DNA can be used.
The extremely detailed regulation of the use of each piece of DNA is through many large and small molecules that trigger, enhance, and stop transcription of DNA code to RNA—called transcription factors. Large proteins called promoters, repressors, and enhancers bind to the DNA, as well as millions of small and large RNA from non-coding regions.
 The next layer of complexity occurs in the messenger RNA, which, by itself, edits the code taken from the DNA. This editing, called alternative RNA splicing, can make up to 500 different proteins from the same pieces of DNA (see post). The RNA cuts out sections and sews others together. Recently, it has been demonstrated that the pieces that are sewn together by the messenger RNA may come from multiple different areas that used to be called individual genes.
The next layer of complexity occurs in the messenger RNA, which, by itself, edits the code taken from the DNA. This editing, called alternative RNA splicing, can make up to 500 different proteins from the same pieces of DNA (see post). The RNA cuts out sections and sews others together. Recently, it has been demonstrated that the pieces that are sewn together by the messenger RNA may come from multiple different areas that used to be called individual genes.
From the messengerRNA, the protein is built into a row of amino acids that forms the protein. The three dimensional folding of this row of amino acids is so complex, that currently, all the supercomputers in the world would take  2,000 years to calculate the folding of one average sized protein (approximately 400 amino acids). Especially neurons have many much larger proteins, each in an exact shape. (see post Protein Shape is Function). We simply cannot know how these many amino acids, each with a complex 3D shape, will fold when put in a row.
2,000 years to calculate the folding of one average sized protein (approximately 400 amino acids). Especially neurons have many much larger proteins, each in an exact shape. (see post Protein Shape is Function). We simply cannot know how these many amino acids, each with a complex 3D shape, will fold when put in a row.
But, somehow, the cell knows what the shape will be and edits the code to form very specific structures. With the help of special chaperone molecules, in a millisecond, the protein folds into primary, secondary, tertiary and then quaternary shapes. The entire functions of the large protein machines are based on shape and the instant folding. (see post on Protein Folding)
This post will attempt to summarize what the new studies tell us about this vast complexity.
DNA History: Genome Study
In 2003, after ten years and great expense, the Human Genome study found only 24 thousand genes in 1.5% of the DNA. This shocked the scientific community because of the small number of so-called genes; previously the dogma was one  gene makes one protein. That would mean we could only have 24 thousand proteins. But, we already knew there are many thousands of different proteins, different in each cell type and each brain region.
gene makes one protein. That would mean we could only have 24 thousand proteins. But, we already knew there are many thousands of different proteins, different in each cell type and each brain region.
The Genome Project showed that 98.5% of the DNA was not making proteins. Also, the scientific community believed that by mapping the “genes” mutations would be identified for many diseases. Ten years after the genome project, almost no disease mutations were found in the genes. Very recently, it has been shown that most mutations that could cause disease must be in the regulatory (“junk”) DNA or the protein transcription factors that regulate the start and stopping of DNA usage.
DNA History: ENCODE in 2012
In 2012, ENCODE, Encylcopedia of DNA Elements, published many papers at once after ten years of work in 160 laboratories around the world. ENCODE found a fantastic amount of active DNA making RNA of various kinds—in at least 20% of the DNA and perhaps 50%. Twenty percent of DNA being active means that the regulatory regions of DNA are at least thirty times larger than all the “genes.” (see posts Encode 1 Mind and Molecular Genetics, Encode 2 New Genetic Landscape, and Encode 3 Encode and Evolution).
(Another fifty percent of the DNA is found to be in the form of jumping genes – virus like pieces of DNA that sew themselves in and out of the genome and copy themselves. These are strands of DNA that can possibly code for whole arms of proteins and are probably, along with viruses, the real drivers of evolution—see post “jumping genes” where the battle to control jumping genes in each cell is described)
 ENCODE showed that much of the previously called “junk” DNA was, in fact, active regulatory DNA making large and small RNAs that perform a fantastic amount of tasks. It found millions of factors that influence DNA. There has been rancorous debates since ENCODE by those who still want to maintain that most of the regulatory DNA is junk—these are often scientists who are committed to a theory that all of this is random.
ENCODE showed that much of the previously called “junk” DNA was, in fact, active regulatory DNA making large and small RNAs that perform a fantastic amount of tasks. It found millions of factors that influence DNA. There has been rancorous debates since ENCODE by those who still want to maintain that most of the regulatory DNA is junk—these are often scientists who are committed to a theory that all of this is random.
ENCODE found 4 million different switches that were active, often operating in multiple places at once; 18,000 places where active RNA is made; and 8 million different particles interacting to regulate these RNAs. Basically, ENCODE found that far from being junk, at least 20% of the DNA is made into important small and large RNA particles that have a use. Since then, many studies have begun to show the importance of these large and small non coding RNA.
DNA History: New RNA Code Discovered in 2013
During the same period, the importance of alternative messenger RNA splicing was described. It was found that the messenger RNA edits and alternatively splices together pieces of DNA from multiple places, including multiple “genes”, and forms as many as  500 proteins from one piece of DNA previously called a “gene’.
500 proteins from one piece of DNA previously called a “gene’.
In 2013, a new RNA code was found superimposed on the other code. This new code consists of a 3D structure of the RNA. Until now, messenger RNA was considered to be a linear molecule representing the letters to make a protein. But, now it is shown that most often RNA will form hairpin loops where regions of RNA have complementary sections of RNA code. This loop folding forms a secondary structure that appears to have significance in translation of RNA code into proteins. The studies showed that even closely related individuals could have different folding structures. 1900 different mutations were identified that affect these RNA structures. The other finding is that when messengerRNA meets the ribosome to present the code that will form the protein, cellular proteins must actively unfold the mRNA.
DNA History: “Duons” Described in 2013
In 2013, a very surprising discovery found that sections of DNA have a dual purpose, comprising two completely different codes at once. These regions were called “duons.”
 The genetic code consists of four letters, three of which are used for each word—words are called codons. Each codon corresponds to one of the twenty-two amino acids, or a stop signal. With four letters and three combined to form codons, there are 64 possible codons. A different number of codons correspond to each amino acid. Some of the amino acids correspond to 6 different codons, and others only one. It turns out that choosing different codons, even for the same amino acid, can have unusual effects.
The genetic code consists of four letters, three of which are used for each word—words are called codons. Each codon corresponds to one of the twenty-two amino acids, or a stop signal. With four letters and three combined to form codons, there are 64 possible codons. A different number of codons correspond to each amino acid. Some of the amino acids correspond to 6 different codons, and others only one. It turns out that choosing different codons, even for the same amino acid, can have unusual effects.
Until very recently, it was assumed that transcription factors bind only in the regulatory regions of DNA next to the “gene”. Transcription factors include the up-regulation of the gene activity—called activators or promoters; enhancement or co-activation of the other factors—called enhancers or co-repressors; and down-regulation—called repressors.
The big new discovery is that transcription factors bind inside at least 13% of “genes” themselves, not just the regulatory regions nearby. This was truly shocking since the DNA in the gene, then, must code for two meanings at once for two entirely different purposes.
The first purpose is to transcribe DNA code to RNA code to make proteins. The second purpose is to bind regulatory factors, as the regulatory regions do to trigger other genes. Duons were found to affect the exact choice of the codon and had an influence the alternative RNA splicing in the messenger RNA at the next step. But, its full function must be worked out.
DNA History: Bacterial DNAs Influence Described 2013
 Each human being has 100 trillion bacteria and 1000 trillion viruses in their gut representing another 3 million active genes producing active products many of which influence human function. The totality of these active genes has been called the hologenome and is, in fact, the real genome of the human being—each microbe with its own unique genetic mechanism. Recently, it was found that the activity of these microbes is as significant as the metabolism of our liver. (See post on hologenome and intestine epithelial cell)
Each human being has 100 trillion bacteria and 1000 trillion viruses in their gut representing another 3 million active genes producing active products many of which influence human function. The totality of these active genes has been called the hologenome and is, in fact, the real genome of the human being—each microbe with its own unique genetic mechanism. Recently, it was found that the activity of these microbes is as significant as the metabolism of our liver. (See post on hologenome and intestine epithelial cell)
DNA History: FANTOM Finds Promoter and Enhancer Complexity in 2014
The new study from FANTOM, in sixteen papers last week, finds greatly increased complexity with promoters and enhancers. FANTOM—Functional Annotation of the Mammalian Genome—describes in great detail thousands of enhancers and promoters that start the use of a piece of DNA. The increased complexity comes from FANTOMs observation that promoters work in many different places for each gene and in different places for each type of human cell. Also, for even more complexity, multiple promoters combine into larger machines, some touching the DNA and some only combining with the other promoters. Multiple structures exist for each gene in different places.
 FANTOM’s two studies looked at 800 different types of human cells. The first study creates an atlas of places where transcription starts and stops for each cell type. The second study described enhancers (proteins that increase the manufacture of particular genes).
FANTOM’s two studies looked at 800 different types of human cells. The first study creates an atlas of places where transcription starts and stops for each cell type. The second study described enhancers (proteins that increase the manufacture of particular genes).
These studies looked a many specific cells in different tissues and found 185,000 promoters and 44,000 enhancers. The ENCODE studies used only a small number of cell types. FANTOM used a wide range of human cells. ENCODE studied much wider phenomenon, but FANTOM explored many of the human cells for the promoter and enhancer regions in much greater detail. They were able to find the beginning of mRNA and then identified regions near, where the upstream promoters lie.
 A very significant FANTOM finding is that there are multiple different places where transcription starts for each “gene”, again undermining the notion of the simple “gene.” It, also, found that all the different cell types start transcription using the DNA at different sites. Along with alternative RNA splicing, the entire concept of “gene” has to be reconsidered.
A very significant FANTOM finding is that there are multiple different places where transcription starts for each “gene”, again undermining the notion of the simple “gene.” It, also, found that all the different cell types start transcription using the DNA at different sites. Along with alternative RNA splicing, the entire concept of “gene” has to be reconsidered.
To find enhancers, they used similar techniques, but the enhancer situation is more complex because some enhancers are transcribed in two directions at once—from the center out in both directions and from both DNA strands. FANTOM noted that the bidirectional types of enhancers are the active important versions. Limitations of the study noted that there are probably a much larger number than was found by these techniques.
Also, most cell types have different varieties of these proteins to express the differences in the types of functions, such as a neurons versus a heart or kidney cells.
FANTOM: Specific Human Tissues
Four hundred different human cells were shown to have different regulatory proteins and different active and inactive genes. Each type also responds differently to the environment with different gene changes and interacts with other cell types. Most genes have more than one start site. The regulation of each is complex and varied.
They also looked at 250 types of cancer cells, whose patterns were noted to be extremely variable.
FANTOM compared findings of the mouse and human, to show evolutionary changes. The conserved areas were similar in both. Only 20% of the genes appear to make cellular scaffolding structures that are common to all cells. These genes appeared to be similar in mouse and human. The most similar were in fibroblasts, chondrocytes in bone, and  adipocytes.
adipocytes.
FANTOM found 80% of the genes are used for the specific functions of the types of cells from different human organs. These genes were very different in humans than mice. In human cells there are a large amount of very different enhancers and promoters and combinations of them. The human that were unique included T cells, macrophages, dendritic cells, blood cells and endothelial cells. This is in line with the very different and rapidly evolved human immune system. A previous post described the remarkably complex and intelligent intestinal endothelial cell, which is very different in humans.
Another conclusion was that since only 128 factors could be studied at once, FANTOMs findings are just the tip of the iceberg of complexity at this level.
Genetic Complexity In Many Studies
While ENCODE and FANTOM are large studies consisting of hundreds of research sites working together around the world, many other isolated  findings, also, continue to show the vast complexity of the genetic machinery.
findings, also, continue to show the vast complexity of the genetic machinery.
Recently, it was shown that exercise effects 10,000 genes and that insulin effects 7 thousand genes in the cell.
Recently, neurexin proteins—critical proteins that hold synapses together—were found to have 2500 variations or more.
Postsynaptic densities in synapses involve a thousand interlocking proteins and these are different in most regions of the brain involving vast genetic changes.
When Thought is Added to the Equation
 Perhaps most unusual are the findings of how mental events trigger vast genetic changes.
Perhaps most unusual are the findings of how mental events trigger vast genetic changes.
Human thought instantly alters large circuits of neurons (see post of neuroplasticity). This occurs through a wide variety of different neuroplasticity mechanisms by triggering specific gene networks and the creation of different complex machinery protein in each different brain region.
Perceptions of loneliness are very significant triggers of gene networks in immune cells causing more inflammation—130 anti inflammatory genes have been shown to lack function and 80 pro inflammatory genes were very active.
Meditation or perception of helping the community, also, triggers hundreds of complex genetic changes that stop inflammation and viruses (see post on Meditation and the Brain).
Therefore, social situations, perceptions and thoughts trigger networks of genes deep inside the cells of the immune system, hormonal systems, body organs and brain. Remarkably, the cell knows exactly the shape of the proteins needed for each of the different neuroplasticity mechanisms that occur simultaneously. Recent posts have described many of these neuroplastic mechanisms that occur simultaneously in large brain circuits, each requiring different complex gene networks. These include:
- Manufacture and placement of more AMPA glutamate receptors
- Calcium surge causing lasting increase in signal called LTP and “memory proteins”.
- Dendrites change structure—size of the head of the dendrite
- Post-synaptic density alters 1000 interlocking large proteins in each brain region
- Change of synapse binding molecules—neurexin, neuroligin
- Pre synaptic neurons substitute neurotransmitters & postsynaptic change receptors Signaling cascades from membrane to nucleus altered
- Changes in axon ion channels alters electrical signal
- Balance of inhibition and stimulation changes
- New micro RNA’s alter process
- Interneurons are altered
- Mitochondria in the dendrite alters the strength of the signal
- Proteins altered in extracellular matrix
- Myosin motors and actin structures altered
- Alterations of climbing fibers in cerebellum
- NMDA glutamate receptors substitute their subunits
- Transport motors are substituted
- Exosomes send information from astrocytes to neurons including pieces of DNA
New Studies Reveal Higher Levels of Genetic Complexity
It is not known how many overlapping codes there are.
 There are multiple factors that determine the opening of the histones. Multiple factors determine which pieces of DNA will be used. A fantastic array of promoters and enhancers and large and small RNAs interact in multiple ways to regulate what pieces of DNA are used. Thousands of different multiple large protein promoters operate in multiple start sites and with multiple proteins either binding to the DNA, or forming large structures by attaching to the other promoters. In some DNA there are two superimposed codes at once. Also, somehow, many different mechanisms are used to repair DNA errors (see post).
There are multiple factors that determine the opening of the histones. Multiple factors determine which pieces of DNA will be used. A fantastic array of promoters and enhancers and large and small RNAs interact in multiple ways to regulate what pieces of DNA are used. Thousands of different multiple large protein promoters operate in multiple start sites and with multiple proteins either binding to the DNA, or forming large structures by attaching to the other promoters. In some DNA there are two superimposed codes at once. Also, somehow, many different mechanisms are used to repair DNA errors (see post).
Eight million factors affect the RNA particles that are made from at least 20% of all DNA (maybe up to 50%). Messenger RNA somehow determines multiple different edits from the same pieces of DNA. Pieces are taken from multiple places, strands are cut out and others sewn together without clear direction. New 3D folding of the RNA also forms a new code.
All of these processes create a code of amino acids that the cell knows will form a very specific very complex protein shape. Proteins only work through exact shapes. Although it would take current supercomputers 2000 years to figure out the folding of one 400 amino acid length protein, proteins are actually folded in primary, secondary, tertiary and quarternary structures in a millisecond, with the help of complex proteins called chaperones. Manufactured proteins then affect all the different neuroplastic mechanisms in a large circuit at once. These proteins, also, affect immune cells throughout the body.
Where is the regulation and control for all of this?
How can anyone say that these overlapping codes involving hundreds of thousands of interacting factors is in any way random?
With mental events triggering these gene networks deep inside of cells, how can anyone say that mind does not influence these genetic processes?