 A thought occurs! Instantly, the signal goes out, DNA is triggered and coding of RNA begins. Pieces of RNA are brought together and edited. Some large RNAs travel to the ribosome manufacturing proteins for microtubules and actin, transporting sacs holding neurotransmitters, building structures to seamlessly merge sacs with the cell membrane releasing the neurotransmitters. Other smaller RNA’s fan out across the cell, regulating every aspect of the process. As the dendrite is built, soon a synapse appears with another neuron. The elaborate synapse structure emerges between the two neurons
A thought occurs! Instantly, the signal goes out, DNA is triggered and coding of RNA begins. Pieces of RNA are brought together and edited. Some large RNAs travel to the ribosome manufacturing proteins for microtubules and actin, transporting sacs holding neurotransmitters, building structures to seamlessly merge sacs with the cell membrane releasing the neurotransmitters. Other smaller RNA’s fan out across the cell, regulating every aspect of the process. As the dendrite is built, soon a synapse appears with another neuron. The elaborate synapse structure emerges between the two neurons
A mental event has triggered an enormous molecular cascade.
Previous posts have shown the extreme complexity in the operations of a neuron, and the self-editing of genetic material that occurs inside cells. The complexity of these processes has been difficult to assess with 20,000 genes identified a decade ago in the genome project. Since then multiple regulatory molecules have been found, including protein transcription factors and small pieces of regulatory RNA. In 2001 it was hard to imagine how 20,000 genes would be enough to manage a human brain since a flea species has 31,000 genes and mice and the plant Arabidopsis have just about the same amount as humans.
Researchers in 2001 who completed the historic Genome Project, the first mapping of the code of a human’s genes, were very surprised at the low number of genes compared to the complexity of the human being, and particularly the complexity of the human brain. Individual cells are somehow able to differentiate into all of the different cell types of the body. And these same cells, responding to signals build the monumental structure of the brain. It was not  at all clear how this complexity could be built from 20,000 genes, each making a protein. Each synapse alone has thousands of different proteins.
at all clear how this complexity could be built from 20,000 genes, each making a protein. Each synapse alone has thousands of different proteins.
Later, it became clear that proteins were not made in one piece but are spliced together from multiple pieces of DNA transcribed into multiple pieces of RNA. With multiple splicings it was found that each gene could make many different variations of a protein.
Where is the direction for such a complex process?
Making Neurons
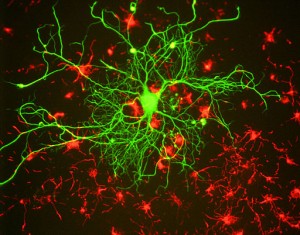 Somewhere in the cell there is a diagram for the vast complexity of the human brain, or, at least, how the cells respond to the environment in making the brain structures. In the last month of the fetus, new brain cells are created and migrate into place at the astonishing rate of 250,000 per minute. This leaves 1 trillion neurons, which are trimmed by experience to approximately 100 billion neurons, the number for the rest of life. Each neuron has on average a thousand connections but can have up to 100,000 dendrites. This creates the astronomical number of synapses of 10 thousand trillion. Each synapse has over a thousand large interacting proteins that vary in the hundreds of different types of neurons (to see these diverse neurons visit the amazing website Neuromorph.org).
Somewhere in the cell there is a diagram for the vast complexity of the human brain, or, at least, how the cells respond to the environment in making the brain structures. In the last month of the fetus, new brain cells are created and migrate into place at the astonishing rate of 250,000 per minute. This leaves 1 trillion neurons, which are trimmed by experience to approximately 100 billion neurons, the number for the rest of life. Each neuron has on average a thousand connections but can have up to 100,000 dendrites. This creates the astronomical number of synapses of 10 thousand trillion. Each synapse has over a thousand large interacting proteins that vary in the hundreds of different types of neurons (to see these diverse neurons visit the amazing website Neuromorph.org).
 There are also ten times as many glial cells (a trillion) that also interact constantly with the synapse. A previous post noted that the glia tap the synapses each night as part of the process of pruning those synapses not being used. Many more synapses are created each day, as well as new brain cells in several places of the brain. This is part of the brain’s ability to change and adapt, the neuroplasticity described in previous posts.
There are also ten times as many glial cells (a trillion) that also interact constantly with the synapse. A previous post noted that the glia tap the synapses each night as part of the process of pruning those synapses not being used. Many more synapses are created each day, as well as new brain cells in several places of the brain. This is part of the brain’s ability to change and adapt, the neuroplasticity described in previous posts.
To further complicate this process the synapses are spatially arranged and operate with different frequencies of pulsed signals.
Crucial Working of DNA
All activity is dependent upon the crucial internal workings of the DNA in the nucleus of the neurons. It is the DNA that instantly responds to make the proteins, which constantly change the shape of the neuron, building new dendrites and synapses.
 To get some idea of the complexity we are about to explore, a previous post describes how cells self edit their own DNA, bringing the error rate in the copying of the DNA during cell division (which causes mutations) from an error 1 in 100,000 down to the astonishing final error rate of only 1 in 10,000,000. This process involves multiple sensors, cutters, joiners, copiers, and editors. Somehow, all of these processes are orchestrated.
To get some idea of the complexity we are about to explore, a previous post describes how cells self edit their own DNA, bringing the error rate in the copying of the DNA during cell division (which causes mutations) from an error 1 in 100,000 down to the astonishing final error rate of only 1 in 10,000,000. This process involves multiple sensors, cutters, joiners, copiers, and editors. Somehow, all of these processes are orchestrated.
For the neuronal processes triggered by thought, the copies of RNA are edited using these same functions: sensors, cutters, joiners, copiers and editors.
The question is how is it all directed?
DNA Regulation
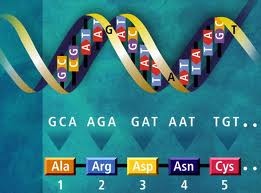 Soon after the double helix structure of DNA was discovered, the vital code was also found. The double helix has two complementary very long DNA molecules that are attracted to each other by small molecular forces. These two hovering molecules form a double spiral in the form of a helix. The long DNA molecule consists of four different nitrogen containing rings, called bases, linked together in a very long chain. These nitrogen rings are guanine, cytosine, adenine, and thymine, known by the letters, G, C, A, T. Any three of these letters code for one of the twenty amino acids that connect together to form the long chain of a protein molecule, or the short chain of a small RNA molecule. There are three million of these letters, or bases, each attracted to their complementary mate in the other helix strand, the two attracted bases together called the base pairs.
Soon after the double helix structure of DNA was discovered, the vital code was also found. The double helix has two complementary very long DNA molecules that are attracted to each other by small molecular forces. These two hovering molecules form a double spiral in the form of a helix. The long DNA molecule consists of four different nitrogen containing rings, called bases, linked together in a very long chain. These nitrogen rings are guanine, cytosine, adenine, and thymine, known by the letters, G, C, A, T. Any three of these letters code for one of the twenty amino acids that connect together to form the long chain of a protein molecule, or the short chain of a small RNA molecule. There are three million of these letters, or bases, each attracted to their complementary mate in the other helix strand, the two attracted bases together called the base pairs.
If DNA in a cell were stretched out it would be 20 miles long. The reason it can be squeezed into the nucleus of a tiny cell is elaborate twisting and looping. The protective twisting is controlled by a set of large proteins; the most important are called histones, the entire structure called chromatin.
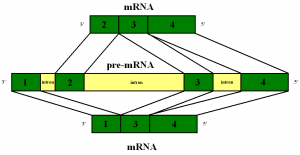 Each gene consists of a long chain of these letters, pieces of which are placed in different regions of the long DNA molecule. Finding the pieces of the DNA that make up the gene, and splicing them together is complicated by the need to first unfold the chromatin. Then, the splicing occurs. Recently, it was found that proteins are not necessarily made in the nucleus, but these spliced RNAs can be transported a long way to dendrites or sites along the axons where RNA ribosomes are waiting (ribosomes are the factories that makes the protein from the RNA strand). Otherwise, all of these proteins would need to be transported on the microtubules to the different regions. These particular RNAs need an address system to arrive at the correct place.
Each gene consists of a long chain of these letters, pieces of which are placed in different regions of the long DNA molecule. Finding the pieces of the DNA that make up the gene, and splicing them together is complicated by the need to first unfold the chromatin. Then, the splicing occurs. Recently, it was found that proteins are not necessarily made in the nucleus, but these spliced RNAs can be transported a long way to dendrites or sites along the axons where RNA ribosomes are waiting (ribosomes are the factories that makes the protein from the RNA strand). Otherwise, all of these proteins would need to be transported on the microtubules to the different regions. These particular RNAs need an address system to arrive at the correct place.
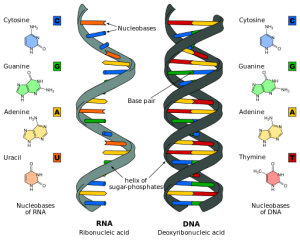
Researchers were further confused to find that the genes were only 1 to 2% of the entire amount of DNA. The rest was considered “junk” DNA. In the past years it became apparent that many newly discovered small pieces of RNA were vital to the regulation process, such as microRNA (in the picture below). These pieces of RNA are not made into proteins and therefore are called non-coding RNA. A previous post noted the increasing number of different vital RNA pieces that have begun to be identified. These have to be coded somewhere in the 98% of “junk” DNA.
New different kinds of regulatory RNAs has mushroomed recently.
Many Types of RNA
 The following is an extraordinary list of recently discovered types of RNA. It is only given to show the complexity just prior to dramatic new developments in the past week, which will be discussed in the next post:
The following is an extraordinary list of recently discovered types of RNA. It is only given to show the complexity just prior to dramatic new developments in the past week, which will be discussed in the next post:
Messenger RNA – the transcript of the DNA letters into RNA code
Ribosome RNA – the large structure that actually builds the protein molecule
Transfer RNA – brings the specific amino acids to the ribosome
Transfer-messenger RNA – rescues stalled ribosomes
Signal Recognition particle RNA – helps membranes
Small nuclear RNA – helps splicing
Small nucleolar RNA – modifies RNAs
SmY RNA – splices messenger RNA
Guide RNA – modifies messenger RNA
Ribonuclease P – modifies transfer RNA
Ribonuclease MRP – helps DNA copy itself
Y RNA – DNA copying
Telemerase RNA – makes telomeres
Antisense RNA – multiple functions in editing
Cis-natural antisense transcript – gene regulation
CRISPR RNA – targets parasite RNA
Long coding RNA – an entire class of RNA’s with multiple functions
microRNA – many different kinds of gene regulation
Piwi-interacting RNA – multiple functions – affects jumping genes
Small interfering RNA – gene regulation
Trans acting siRNA – gene regulation
Repeat associated siRNA – jumping gene regulation
7SK RNA – regulation of signal pathways
Retrotransposon RNA – self propogating parasite
All of these RNAs must come from the non-coding parts of the genome, which were called junk until last week. This was the state of complexity just before the latest breakthrough – ENCODE.
The Encyclopedia of DNA Elements – ENCODE
For the last ten years since the genome project mapped out the 20,000 genes it has not been at all clear how this much regulation can occur. Just after the completion of the Genome Project, an equally profound study began called ENCODE or the Encyclopedia of DNA elements. This vast study, involving many hundreds of research projects around the world, has just made its first set of major conclusions after ten years.
These were published last week in 30 articles at once in the world’s major scientific journals, including 6 summary articles in Nature and one in Science. ENCODE is the largest example of international cooperation in a science project.
From ENCODE, it is now clear the complexity of the human being, including the brain, is accomplished in a vast regulation network in the region of the DNA, once called junk. The complexity is not related to the number of genes. The new research describes a genetic landscape that is almost incomprehensible. The next several posts will attempt to summarize where we are now, after last weeks explosion of data, in understanding the complexity of the neuron and how this might be triggered by mental activity.

