The shape is what determines the functions of proteins, either as a structural element in the neuron, or as an enzyme in reactions. To perform as an enzyme, the protein must have very exact structures, allowing specific molecules to interact with it while encouraging chemical reactions.
Somehow the regulatory network of the cell knows how the protein will fold from the linear code of amino acids. How the code is translated into a three-dimensional structure is unknown. In fact, there are an almost infinite number of different ways the amino acids can fold incorrectly and disastrously for the cell.
For most proteins there appears to be only one or two correct shapes. Many use two shapes for different actions, such as hemoglobin with one shape for oxygen and another for carbon dioxide. Recently a few rare loosely folded proteins have been observed to function.
Protein Shape Determines Function
Shape determines everything in the cell.
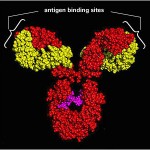 Special proteins are manufactured by the immune system called Antibodies that are shaped to latch onto the surface of microbes. Antibodies are made when special blood cells self edit their DNA to create uniquely shaped proteins.
Special proteins are manufactured by the immune system called Antibodies that are shaped to latch onto the surface of microbes. Antibodies are made when special blood cells self edit their DNA to create uniquely shaped proteins.
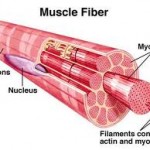 Contractile proteins are shaped to slide over one another for movement such as in muscles, and microbe cilia.
Contractile proteins are shaped to slide over one another for movement such as in muscles, and microbe cilia.
Enzymes are complex proteins that facilitate chemical reactions  by specially binding to the two chemicals, creating a unique space to speed up the reaction. These reactions usually would not have the energy characteristics to occur without the enzyme. Another enzyme version breaks down other molecules, such as food particles in the gut.
by specially binding to the two chemicals, creating a unique space to speed up the reaction. These reactions usually would not have the energy characteristics to occur without the enzyme. Another enzyme version breaks down other molecules, such as food particles in the gut.
 Hormones are proteins that signal complex functions throughout the body
Hormones are proteins that signal complex functions throughout the body
Receptors in membranes are proteins that are triggered when a specifically shaped molecule forms a lock and key.
 Proteins that form Membrane Channels allow electricity to flow in the axon (see post on Electricity in the Brain).
Proteins that form Membrane Channels allow electricity to flow in the axon (see post on Electricity in the Brain).
Structural Proteins like microtubules build the highways of cells, where material is transported (see post The Remarkable Microtubule). They are also stringy and fibrous building cellular structures.
 A Storage Protein provides amino acid for food, such as egg whites.
A Storage Protein provides amino acid for food, such as egg whites.
Transport proteins, like hemoglobin, carry important molecules throughout the body. In photosynthesis, they allow the transfer to electrons to make energy from light.
Protein Folding
Protein folding, when abnormal, causes many important diseases. The prion, an abnormally folded protein, causes other proteins in its path to alter their shape, eventually damaging brains in the process. The folding principles of the prion disease Mad Cow (known also as Creutzfeldt-Jakob disease) are unknown. Recently, there is evidence that other major degenerative neurological diseases may also be related to protein folding, including Alzheimer’s, Parkinson’s, Huntington’s, and amyotrophic lateral sclerosis.
In a previous post on jumping genes, it was noted that entire sequences of DNA representing the shape of sections of proteins are copied in jumping genes, as well as viruses. This commerce in protein sections may be the major driver of evolution.
 Another post noted the unusual fact that cells self edit their own DNA and are able to understand and fix errors in the DNA copying process. A similar editing process occurs when RNA revises the material it takes from the genes before using to manufacture a protein. Another similar editing process occurs with jumping genes, which edit themselves in and out of locations in the gene.
Another post noted the unusual fact that cells self edit their own DNA and are able to understand and fix errors in the DNA copying process. A similar editing process occurs when RNA revises the material it takes from the genes before using to manufacture a protein. Another similar editing process occurs with jumping genes, which edit themselves in and out of locations in the gene.
All of these elaborate and deliberate processes appear to involve cellular cognitive processes. Another layer of this cognitive and coding puzzle is the immensely complicated determination of how the chain of amino acids translates into the very specific all-important three-dimensional shape of the protein. The folding seems to occur immediately, which adds to the mystery.
To date, we do not know enough to translate a sequence of amino acids, which are determined by the DNA sequence, into the all important shape of the protein.
Protein Structure
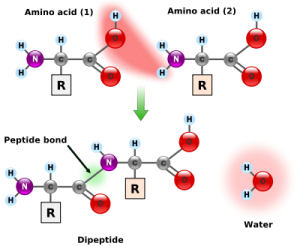 Amino acids are small molecules that consist of a carbon, a nitrogen, an oxygen and unique side chain. When amino acids combine end to end they form a structure with the carbon and nitrogen bond forming a spine like structure that can attach hundreds or thousands of amino acids in a row.
Amino acids are small molecules that consist of a carbon, a nitrogen, an oxygen and unique side chain. When amino acids combine end to end they form a structure with the carbon and nitrogen bond forming a spine like structure that can attach hundreds or thousands of amino acids in a row.
There are twenty different amino acids used in proteins, each with a different side chain. The shape and chemical properties of the 20 side chains are distinct and determine the chemical properties of the specific amino acid (A.A.) and the folding of the long chain. There are actually 150 different A.A.s found in a cell but only 20 are used for proteins.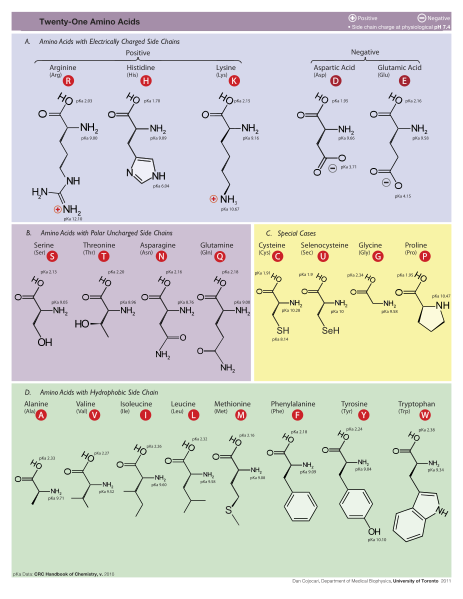
The average human protein is under 400 A.A.s long, but they can be considerably longer with the sarcomere muscle made of 27,000 A.A. in a row. Below is a picture from the Protein Data Base of many proteins to see that there is great range of relative sizes. This can be downloaded for free. 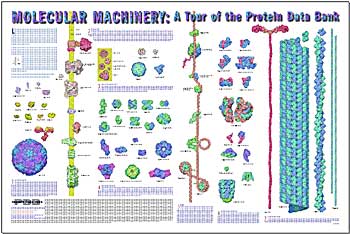 .
.
A.A.s are strung together by what is called a peptide bond, where a carboxyl group, a carbon and oxygen on one end, combines with the amino group, the nitrogen, of another and forms the peptide bond. (see picture). When all the amino acids are strung together forming the protein one end of the entire protein is a carboxyl group and the other end is nitrogen. A small string of A.A.s is called a polypeptide.
When all the amino acids are strung together forming the protein one end of the entire protein is a carboxyl group and the other end is nitrogen. A small string of A.A.s is called a polypeptide.
Four Levels of Folding
Primary Structure:- The sequence of A.A.s is called the primary structure, directly based upon the sequence of DNA, then RNA that produces the code to string together the specific set of A.A.s.
 Secondary Structure – When small local sections of the amino acids start to fold they can form specific local substructures called the secondary structure.
Secondary Structure – When small local sections of the amino acids start to fold they can form specific local substructures called the secondary structure.
Two common secondary structures are the Alpha Helix and Beta Sheets. The alpha helix is the most common, the most predictable from the sequence, and the most regularly shaped structure.
The beta sheet is another regular structure, slightly less common than the alpha helix. These beta sheets can form aggregates called amyloid, a primary finding in Alzheimer’s disorder and other diseases.
Tertiary Structure – This is the three-dimensional structure after folding is complete. The structure is locked into place with specific chemical interactions between A.A.s on the side chains that are located near each other in the final fold.
Quaternary Sructure – Large proteins are often the subunits of a much larger structure made of multiple pieces. (Dimer if two subunits, trimer for three, tetramer for four) Examples of these large structures include many of the membrane channels, the histone molecules, the postsynaptic cleft and many other structures. The same chemical forces stabilize these as the tertiary structure.
Forces Determine Folding – Final Shape
There are a variety of forces operating on the folding protein that determine small local attractions between side chains. These forces do not create the strong co-valent bond that is the basis of atoms forming molecules, but rather are weaker forces that attract and repel loose sections of the molecule. (The peptide bond that links A.A.s in a chain is a strong co-valent bond.)
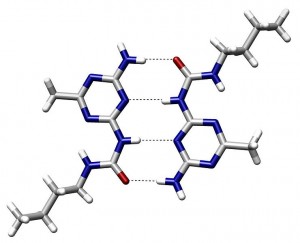 Hydrogen bonds – This bond occurs when there is electromagnetic attraction of a positively charged hydrogen atom and a negatively charged atom, such as nitrogen, oxygen, or fluorine. This can occur in many molecules large and small, such as water and DNA. Since all cellular molecules are surrounded by water, these hydrogen bonds are very frequent.
Hydrogen bonds – This bond occurs when there is electromagnetic attraction of a positively charged hydrogen atom and a negatively charged atom, such as nitrogen, oxygen, or fluorine. This can occur in many molecules large and small, such as water and DNA. Since all cellular molecules are surrounded by water, these hydrogen bonds are very frequent.
Van der Waals interactions – These are close range reactions from a variety of sources that influence solids and liquids and affect proteins when they are tightly packed.
 Backbone angle preferences – These are preferred angles of bonding along the spine of the protein.
Backbone angle preferences – These are preferred angles of bonding along the spine of the protein.
Electrostatic interactions – Amino acids can have other positive and negative charges and attract and repel each other.
Hydrophobic Interactions – Hydrophobic means “water fearing.” Molecules can be polar meaning they have slightly positive and negative charges on either side, or non-polar where there is no charge. Since water is a polar molecule and all of the cellular reactions occur in water, the non-polar molecules, such as fat or oil, will accumulate in a clump and will not mix with the water. This is seen when you attempt to mix oil and water.
 Likewise, sections of the proteins that are hydrophobic tend for form a clump on the inside of the protein, and the charged polar molecules are on the surface interacting with the water that surrounds it.
Likewise, sections of the proteins that are hydrophobic tend for form a clump on the inside of the protein, and the charged polar molecules are on the surface interacting with the water that surrounds it.
Chain Entropy – This is a concept from thermodynamics where a protein will go to its lowest energy state, which is the final stable folding shape.
When proteins fold correctly, they end up in a state with low energy that is a very stable state. If they start folding in a wrong way first, the entire folding process can be disturbed. These intermediate partially folded states each have an energy state. Scientifically this is visualized as a funnel energy graph where the high energies are partial or incorrectly folded proteins and down at the bottom of the funnel is the only way the chain of amino acids can really be at rest. Theoretically this funnel could be drawn for each protein, but in reality this information is not known for most proteins because it is too complex.
The Folding Pathway
When you look at a relatively small protein of 100 Amino acids, the folding can occur in an almost infinite number of ways. Starting with the very first twist, it can fold in many directions. Each further twist adds an enormous number of possibilities.
To understand this number of possibilities, it must be considered that with only a 100 amino acids the number of possible folds is an extremely large number. If 100 billion fold attempts were done each second, then the number of possibilities would be greater than the age of the universe, approximately 10 billion years. These possibilities are impossible for the largest computers to consider.
In fact, the average human protein is 370 A.A.s, a much larger number of possibilities.
Proteins first appear to fold in the secondary structure. As the folding proceeds it becomes more stable. First local structures occur such as helixes and specific turns, then 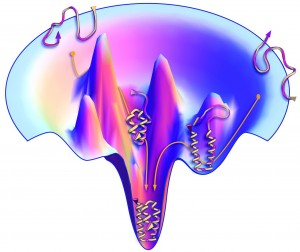 larger structures occur. Studies have shown that there are many possible folds of a protein that have high energy because they are not stable. In other words they will eventually fold into the more permanent shape, which has a stable low energy.
larger structures occur. Studies have shown that there are many possible folds of a protein that have high energy because they are not stable. In other words they will eventually fold into the more permanent shape, which has a stable low energy.
This landscape of energy is shown in the funnel diagram (left.) At the bottom of the funnel is the resting state with the lowest energy, and the most permanent shape. Higher in the funnel are shapes that can still change and therefore have more energy. The picture shows that only one, or at least very few, possible folds out of an infinite number are stable.
Some Results for Small Proteins
Extensive research for fifty years has lead to some results, but only in some very small proteins.
Small molecules can be calculated to some degree but not their thermodynamic or energy characteristics. Recently, a group was able to make observations of specific alpha and beta structures, and several rules were deduced. Thus in a small group of structures involving alpha and beta sheets, new structures were recently created with these rules.
 The picture to the right shows a calculated shape of several small proteins (red), and the actual observed shape (blue).
The picture to the right shows a calculated shape of several small proteins (red), and the actual observed shape (blue).
Anything larger cannot be calculated. At first it was thought that certain amino acid structures would fold in certain ways, but that has been shown to not be true.
Large Scale Calculation of Protein Folding
The protein data bank currently has analyzed 80,000 structures with the details of each atom. These represent 4,000 different structural families of proteins with 1200 folds. The downloadable free picture shown above previously is from the data bank and shows the extremely varied sizes and shapes of proteins.
Even knowing these exact final shapes from Xray observation the folding cannot be deduced. Some progress has occurred with less than 100 amino acids, but not consistently.
There are a number of attempts to find the folding patterns of proteins with known shape. These use unique networks of a large number of individual computers working together. One such attempt is a yearly public competition to describe an algorithm for a particular known protein. This competition has occurred for eight years.
One project called Folding@home has more than a million registered users where local computers are used with an average of a quarter million computers working at any moment on calculating folding. Another computer game Foldit, computer gamers compete to calculate folding.
Both computing power and knowledge of force fields are increasing, but still the numbers are overwhelming.
While a huge amount of research continues, there is still little known. A recent lead summary article in Science summarized the state of knowledge about protein folding. Science currently CANNOT:
Predict the shape of an amino acid sequence
Describe the energy landscapes of folding
Observe the details of folding
Devise algorithms to predict binding to proteins
Understand how such dense molecules can stay intact in cells
Understand the folding diseases
How Can They Fold So Fast: Chaperones
 One thing we do know is that are large proteins called chaperones assisting other large molecules in proper folding, including other proteins. When the first chains of peptides (strings of amino acids – see picture at left) are formed they might have a tendency to fold in a way that will not help the total protein folding. In the crowded cell it would be easy for side chains to stick to other side chains. One of the major functions of chaperones is stopping small polypeptides from folding prematurely. To perform this function they must not
One thing we do know is that are large proteins called chaperones assisting other large molecules in proper folding, including other proteins. When the first chains of peptides (strings of amino acids – see picture at left) are formed they might have a tendency to fold in a way that will not help the total protein folding. In the crowded cell it would be easy for side chains to stick to other side chains. One of the major functions of chaperones is stopping small polypeptides from folding prematurely. To perform this function they must not 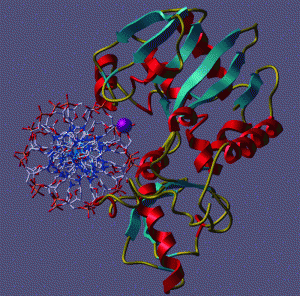 bind too tightly or too loosely, so the protein is protected but not bound.
bind too tightly or too loosely, so the protein is protected but not bound.
Some chaperones help the histones that surround and protect DNA to fold and unfold correctly. Many chaperones are part of a group of proteins called Heat Shock proteins. These are molecules that are manufactured when there is stress, often increased temperature, to help molecules not be influenced by the increased heat to fold incorrectly. Incorrect damaging folding, called denaturing can be observed when we cook an egg and see the proteins become the “whites” of the egg.
How Can They Fold So Fast: Quantum
Folding occurs in microseconds. The assumption in the field of protein folding, as with all molecular science, is that the process is random and blind, and that the folding gropes through random attempts to follow the energy states of each fold.
But, this could not explain the speed at which proteins fold. And biological reactions often go to a higher energy state in order to reach a lower state eventually.
A quantum answer would not be random and blind. It could find the best, lowest energy, by quantum superposition (see post Quantum Effects in Life).
 A previous post on quantum effects in biology showed that the electron in photosynthesis instantly finds the most efficient exact path to the center. In the same way, the protein strand of amino acids seems to know which of the almost infinite possible folds it should do instantly.
A previous post on quantum effects in biology showed that the electron in photosynthesis instantly finds the most efficient exact path to the center. In the same way, the protein strand of amino acids seems to know which of the almost infinite possible folds it should do instantly.
Is protein folding an even larger example of quantum effects in biology? Recent research seems to lead in this direction.
There are also other very unusual findings. While it is known that proteins have a very specific shape, or several different shapes, it fact there are some proteins recently discovered that have a much looser shape, or a vibrating shape and they also function. These could be other forms of quantum superposition.
Mind Works Through Regulatory Network
Posts on ENCODE have shown that cells self edit their own DNA, and the RNA’s edit the transcript, directed by millions of regulatory factors. Discussion of jumping genes showed that the most probable mechanism of evolution is not point mutations, but rather the copying of entire strands (shapes) of DNA, which are then modified and used for innovations. A question was raised about cellular cognitive function in self-editing, and in the innovations of jumping genes.
 The effects of editing DNA or RNA strands changes the shape of the protein, this shape known only after full folding of the protein. This folding of the protein is not currently predictable but it must be based upon the amino acid sequence. Somehow the editing of the genetic sequence is translated into the 3-D spatial characteristics of the many different proteins.
The effects of editing DNA or RNA strands changes the shape of the protein, this shape known only after full folding of the protein. This folding of the protein is not currently predictable but it must be based upon the amino acid sequence. Somehow the editing of the genetic sequence is translated into the 3-D spatial characteristics of the many different proteins.
Somehow, the cell knows the shape that will come from the sequence. The most advanced computers can’t do it, but the cell is able to self-edit.
The mind works somehow through this genetic regulatory maze in neurons to determine the specific code, translated by unknown measures, into the three dimensional structures and functions in neurons.
Next week we will continue with a look at some of the very specific spatial characteristics of proteins in neurons and how this might be activated by the mind.
