 Alternative splicing of messenger RNA has been shown to be critical for the development of the human brain. The ability to make many new and complex proteins allowed the development of the enormous molecular complexity in different neurons and in different regions. For some reason, in evolution humans developed the ability to use alternative splicing much more than other species. This ability is most prominent in the brain. A previous post two years ago described this phenomena, but recently, the mechanisms have been worked out. This post updates the most recent understanding of how the human brain can make vastly more and complex proteins than other organs and other species.
Alternative splicing of messenger RNA has been shown to be critical for the development of the human brain. The ability to make many new and complex proteins allowed the development of the enormous molecular complexity in different neurons and in different regions. For some reason, in evolution humans developed the ability to use alternative splicing much more than other species. This ability is most prominent in the brain. A previous post two years ago described this phenomena, but recently, the mechanisms have been worked out. This post updates the most recent understanding of how the human brain can make vastly more and complex proteins than other organs and other species.
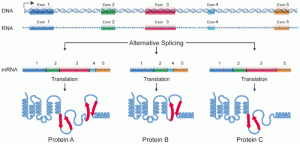 DNA codes for messenger RNAs, which codes for proteins that are made in ribosomes. It was once believed to be simple, but in fact, it is not. Less than 2 percent of the total DNA makes proteins. A great amount of DNA (at least 20 times the amount that codes for proteins) makes small and large RNAs that have regulatory functions in the vast genetic complexity. No one knows how all of this can be regulated or organized.
DNA codes for messenger RNAs, which codes for proteins that are made in ribosomes. It was once believed to be simple, but in fact, it is not. Less than 2 percent of the total DNA makes proteins. A great amount of DNA (at least 20 times the amount that codes for proteins) makes small and large RNAs that have regulatory functions in the vast genetic complexity. No one knows how all of this can be regulated or organized.
Previous posts have described the general concept of alternative splicing. Another post described the unique ways that these RNAs have influenced brain evolution. Other posts have described vast genetic complexity. First, there is a very brief summary of information from the previous posts about alternative splicing.
Brief Summary of Alternative Splicing
 The early simplistic notion was one particular region of DNA, called a gene, makes one protein. The genome project found that there were too few “genes” to make all the proteins. It was found that when making a protein, some of the DNA material is edited out in a very complex process (introns edited out, exons kept). Later, it was noted that an even more complex process occurs where exons themselves are edited by being cut, reorganized and pasted.
The early simplistic notion was one particular region of DNA, called a gene, makes one protein. The genome project found that there were too few “genes” to make all the proteins. It was found that when making a protein, some of the DNA material is edited out in a very complex process (introns edited out, exons kept). Later, it was noted that an even more complex process occurs where exons themselves are edited by being cut, reorganized and pasted.
Even more surprising, some of the DNA that is gathered for one messenger RNA is taken from multiple different so-called “genes”. The critical large study for these findings is ENCODE, which was completed ten years after the Genome project (ENCODE = encyclopedia of DNA elements). Please see posts on ENCODE for details Mind and Molecular Genetics of the Neuron Part 1, Part 2, and Part 3. Because of this the basic definition of “gene” will have to be changed.
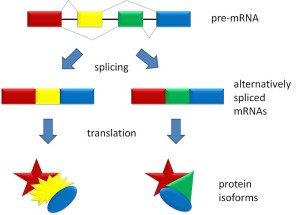
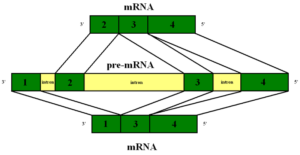
Before the messenger RNA is finalized, a “pre-messenger RNA” (pre-mRNA) is formed which includes sections of DNA that will be used for the protein code, called exons, and usually longer sections that will be edited out, called introns. There can be as many as 100 different introns and exons in a pre messenger RNA strand.
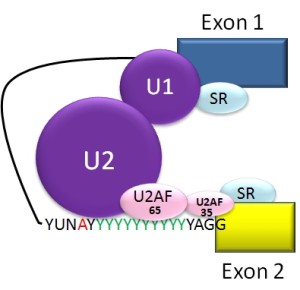
An elaborate molecular machine, called the spiceosome, accomplishes the critical editing. The spliceosome is composed of multiple pieces of small nuclear RNA (snRNA) named U1, U2, U4, U5, and U6, with accompanying proteins. It is this very large complex machinery made from multiple RNA pieces and proteins that does the actual splicing.
Greatly complicating this process, in different types of cells the very same sequence of DNA can be considered an exon or an intron, creating a tremendous variation in the construction of proteins between different cells and different tissues. Also, the point at which the cut is made is not as fixed as it seemed at first. Therefore, it has been found that specific types of cells, such as those from organs like the brain, the kidney, etc., have multiple alternative splicing patterns.
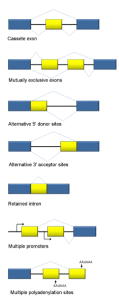 There are many different types of alternative messenger RNA splicing; some types have been clearly defined, but continued research seems to come up with more and more different kinds. This is also complicated by the fact that there can be multiple different steps in the process.
There are many different types of alternative messenger RNA splicing; some types have been clearly defined, but continued research seems to come up with more and more different kinds. This is also complicated by the fact that there can be multiple different steps in the process.
- The first type involves introns being cut out.
- A second involves an exon (usually considered a piece that will stay) being spliced out.
- Another involves two exons in a series, with only one of the two being kept in an alternating pattern.
- There are a number of types of alternative splicing patterns where different parts of the intron are cut. Sometimes, mutations near the edge of the intron can affect this type of process.
- Another type is where the intron is retained.
Somehow, the cell knows exactly what the shape of the new protein will be and how it will fit with the thousands of other exact shapes of proteins in the neuron.
Our greatest supercomputer cannot calculate the folding of an averaged sized protein. It would take a thousand years of computation for one to be deciphered. Therefore, it is not at all clear how the cell knows exactly what new proteins will fit with new brain development and fold them in a millisecond.
Humans use alternative splicing much more than other cells and for many very specific functions. But, of all cells, neurons have the most complex alternative splicing making a host of very unique proteins in all brain regions and neuron types. Alternative splicing, it turns out, is a very important factor making the human brain unique.
Unique Place of RNAs in Brain Evolution
 Alternative splicing is very different in different animal species and much more complex in humans and other more intelligent animals. In humans, 95% of the time there is alterative splicing if the messenger RNA is made from several different gene sources.
Alternative splicing is very different in different animal species and much more complex in humans and other more intelligent animals. In humans, 95% of the time there is alterative splicing if the messenger RNA is made from several different gene sources.
Alternative splicing is a critical source of evolutionary change differentiating primates and humans from other creatures such as worms and flies with a similar number of genes. Alternative splicing is very different in different species. Specifically primates, and especially humans have by far the most complex alternative splicing.
One important example of alternative splicing are kinases, which perform phosphorylation, that is, adding high-energy phosphorus onto a molecule—a process critical to many important signaling channels in cells, and especially neurons. Kinases are the largest and most critical protein family for all types of cellular functions.
Research shows increasing amounts of small and large RNAs that do not make proteins and are from regions in the DNA that were considered “junk”. The ENCODE research showed a great amount of this non protein coding RNA does in fact make critical RNAs, small and large, that perform regulatory functions of all types. These have, also, been critical in evolution, with more complexity in humans. See posts on intelligent small and large RNAs for more information on this topic.
Making New Neurons
The alternative splicing in neurons is not only more common than other cells, the mechanisms specifically have not changed much in recent human evolution—this is referred to as being “highly conserved.” Conserved genes are the most critically important genes, which are most protected from alterations. Alternative splicing is now known to be extremely important in the neuron for the creation of neuron networks in the fetus, and for specific complex interactions among groups of proteins. An example of a large group of proteins occurs in the postsynaptic density (PSD) where up to a thousand different proteins create the landing dock for the neurotransmitters effect. PSDs are quite different in different brain regions and neuron types.
When new neurons are made from stem cells, unique gene networks are triggered that transform the new cell into a particular type of neuron. Different networks of genes are then involved in the back and forth communication between that cell and other cells to allow the neuron to travel to its special place; to create very exact axons, dendrites and synapses; and to be able to respond to changes with neuroplasticity. Specific new proteins are created for these purposes by alternate splicing. Specific tissues engage in particular types of editing. There are many complex proteins helping the editing process and aberrations in these neuron proteins can cause neurological disease.
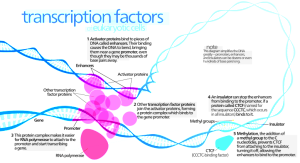
The actions of the regulatory protein products acting on DNA are complex. They are discussed as acting relatively nearby (cis) or in far away places (trans) from the location of their gene production. It has been easier to study the close by cis regulation, but trans research is now advancing. Trans research includes large RNAs that have affects over multiple regions of the genome.
In neurons there are particularly complex groups of molecules that interact to determine the exact type of editing that goes on. They are called splicing enhancers or silencers. Attempts are being made to define a “splicing code” that is discussed in a previous post, where it can be predicted whether alternative splicing will go on. There is now better understanding of what DNA structures will be elaborately spliced. For example, cis regulation usually occurs within 300 DNA base pairs. These are very highly conserved and regulated. In fact, several of these complex regulatory complexes had a rapid evolution along with the rapid evolution of the human brain.
Splicing Regulatory Networks
Each gene consists of the usable exons (and the introns that are edited out). A sub set of these exons becomes a particular network for elaborate regulation. The description of these regulatory gene networks has been the greatest research advances in the past several years.
One regulatory network involves genes that are associated with synapse creation and the guidance of the axon (Nova1 and 2).
Another set of regulatory proteins assist alternative splicing during brain development. They are vital for the development of the cerebellum and avoidance of seizures. These different proteins (RBfox1, 2 and 3) are only in particular spatial regions of the developing brain. The regulation of the alternative splicing in these different regions determines the development of the particular brain regions.
Another set of regulatory proteins (Ptbp1, 2) act by stopping alternative splicing in most regions, but do the opposite in other very specific regions. These mechanisms inhibit particular exons from being used as part of the alternative splicing. An example is when these particular genes function strongly in stem cells, but are inhibited as soon as the stem cell becomes a neuron. Specific exons are not used to make particular alternative proteins, but allow many other alterative spliced new proteins. The alternative splicing is therefore critical to the different stages of brain development.
 Another set of regulatory proteins that modify alternative splicing are nSR100 and sRRM4. These occur in sensory regions as well as many other sub regions. These regulatory factors influence alternative splicing in the creation of a host of proteins that affect the microtubule scaffolding.
Another set of regulatory proteins that modify alternative splicing are nSR100 and sRRM4. These occur in sensory regions as well as many other sub regions. These regulatory factors influence alternative splicing in the creation of a host of proteins that affect the microtubule scaffolding.
Previous posts noted that these microtubules respond immediately to thought by altering structures for neuroplasticity. They are, also, critical in membranes at the synapse. These regulatory factors stimulate use of particular exons that are needed for axons to grow, to build the cortex structure and to drive the nerves of the diaphragm.
Many different splicing factors add together to give particular organs their particular functions. The various factors mentioned above operate together to create many different unusual proteins from the exons that they regulate. This coordination occurs in both cis (nearby) effects as well as trans (effecting genes at great distance). These patterns of multiple interacting splicing factors are very important in determining the unique characteristics of individual neurons. Examples have been found whereby these factors determine the unique properties of GABA neurons and cholinergic neurons.
Neuronal Functions
Exons that are used to make specific proteins in the brain tend to be part of extensive interacting protein networks. They become hubs in the gene networks and interact widely causing there to be more alternative splicing. A very unusual finding is that micro exons (very small exons) are particularly likely to be used in neurons. Somehow, interactions of the various splicing regulators more often influence micro exons. Certain brain diseases, such as autism, particularly have problems with micro exons.
Micro exons appear to be very important in switches of function, such as occurs when a stem cell makes a particular type of neuron. They are particularly important in neuronal signaling. A change in micro exons can change a protein from a repressor to an activator. Several micro exons affect transcription factors critical for the development of the brain. A set of micro exons can form a network that affects a particular network of genes in the neuron. These can be very powerful switches.
There are even more complex regulatory events that are involved with neuronal alterative splicing. One is called NMD (nonsense-mediated mRNA decay) and is a pathway that observes errors in using genes. It is one of many error correction pathways (described in the post Neurons edit their Genes). They eliminate certain parts of messenger RNA that have mistaken exons. In fact, regulatory factors mentioned above work with this NMD pathway in very complex ways.
Another function of these alternative splicing regulatory networks is that it, also, regulates how many proteins will be produced. One mechanism used in this is to keep introns that cause destruction of the transcript. This mechanism is, also, highly regulated by the interacting regulatory systems.
One of the most documented alternative splicing regulatory networks involves neurexin, a critical molecule holding together synapses. This network of splices creates many different proteins that form a wide variety of different synapses. Neurexins (pre synaptic) and neuroligns (post synaptic) are proteins that connect and hold the pre and postsynaptic cells together for a synapse. Neurexin has two genes making six major different neurexins. The alternative splicing is highly regulated at special splice sites. In fact, there are thousands of different neurexins made in this alternative splicing process used in different brain regions—2000 have been identified as different alternate splices. These different proteins create different types of synapses throughout the brain.
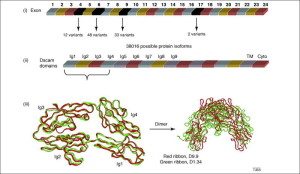
Another critical receptor on the surface of neurons is Dscam1. For this molecule 38,000 different splice variations have been calculated in the fly nervous system. The Dscam1 gene has four different groups of alterative exons as well as other variable regions. This large number of variants has been found to be critical in different brain circuits. A recent study found 18,000 active variations that occur independently.
A similar system in the human brain called Pcdha (clustered protocadherins) produces enough variants so that thousands of neurons can each have individual identities in circuits. There are many other interacting networks involved with these that are just being discovered.
New technology is constantly being applied to these studies. Recently, the very exciting CRISPR techniques, where specific exons can be deleted very accurately, are being applied to this important area. Cis effects have been easier to determine and research is now ongoing for trans effects at a long distance. A previous post described the new frontier of long non-coding RNAs (lncRNAs) that will provide information of these major trans effects from very large active regulatory RNAs. Some of these have been shown to be influential in alternative splicing regulation.
Vast Complexity of Alternative Splicing in Neurons
 How can any reasonable person think that this is a random process? How can anyone think that this process has no direction? The problem is finding where the direction comes from. Clearly not from the DNA, or RNAs which are being manipulated in the process. The circular logic includes proteins uniquely formed that regulate the very process making more elaborate proteins from alternative splicing. There has to be some other source of direction for all of this.
How can any reasonable person think that this is a random process? How can anyone think that this process has no direction? The problem is finding where the direction comes from. Clearly not from the DNA, or RNAs which are being manipulated in the process. The circular logic includes proteins uniquely formed that regulate the very process making more elaborate proteins from alternative splicing. There has to be some other source of direction for all of this.
There are now quite a number of interlocking systems of hierarchal regulation of the genetic systems—many different epigenetic markings on DNA and histones and large numbers of proteins and  other factors that stimulate and repress DNA actions. But several of these processes are completely beyond even the most advanced supercomputers’ ability to compute. This includes millions of regulatory particles discovered by ENCODE. Protein folding cannot be calculated with our most powerful supercomputers, but occurs in milliseconds (see post). The folding is exactly regulated since the shape determines all actions. Somehow thousands of precisely shaped proteins are produced through alternative splicing.
other factors that stimulate and repress DNA actions. But several of these processes are completely beyond even the most advanced supercomputers’ ability to compute. This includes millions of regulatory particles discovered by ENCODE. Protein folding cannot be calculated with our most powerful supercomputers, but occurs in milliseconds (see post). The folding is exactly regulated since the shape determines all actions. Somehow thousands of precisely shaped proteins are produced through alternative splicing.
Splicing mechanisms, somehow, know exactly what the new shape will be for the exact function. Alternative splicing makes sure that the new shapes are constantly evolving for a better, more complex brain. Where is the direction for all of this editing of precise protein shapes?
Can there be any answer other than mind or consciousness directing the molecules, organelles, cells, organs and creatures.